The ability to rapidly and effectively evaluate the performance of customized next-generation sequencing (NGS) panels is critical to provide high-quality sequencing solutions to customers. New England Biolabs®, together with Directed Genomics®, is developing a new offering, NEBNext Direct® Custom Ready Panels, which will allow researchers to select from a large library of genes for which baits have been developed and optimized, thus enabling rapid deployment of customized target-enrichment panels. Directed Genomics has been collaborating with SeraCare Life Sciences in order to streamline the optimization and characterization of NEBNext Direct target enrichment panels.
NEBNext Direct is a novel target-enrichment method; it is hybridization based, but it differs from standard hybridization-based methods in that the genomic DNA capture is performed first, without preliminary preparation of an NGS library prior to capture. The enriched material is what is then put into the library preparation protocol. This approach results in improved uniformity of coverage and the ability to incorporate unique molecular indexing.
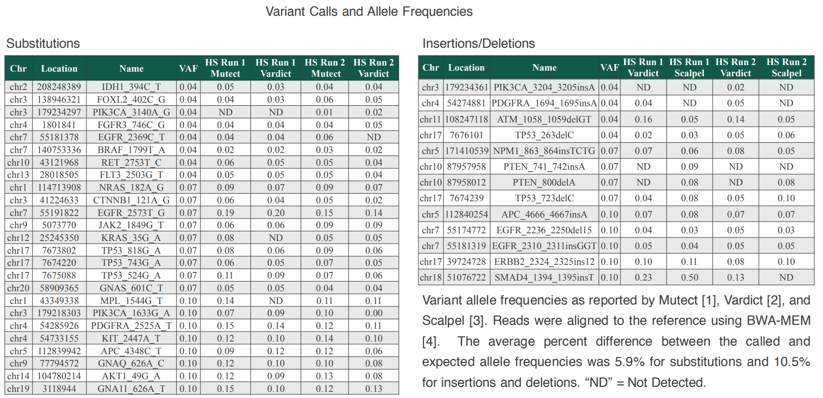
A proof of concept study showing “Reliable Interpretation of NGS Data Using Well-Established, Highly Multiplexed Reference Materials” was presented at the recent Association for Molecular Pathology (AMP) annual meeting. In the study, SeraCare provided the Seraseq™ Tri-Level Tumor Mutation Mix v2, which is a reference material with 40 variants across 28 cancer-relevant genes at variant allele frequencies (VAFs) of 4%, 7%, and 10%. Two panels were developed to target these variants. One panel was a small, targeted “hotspot” panel capturing 4.5 kb of sequence, and the other was a more comprehensive panel capturing 97 kb of sequence. The Seraseq Tri-Level Tumor Mutation Mix v2 fragments were captured using the NEBNext Direct technology and subjected to NGS, and then analyzed using open source variant callers, such as MuTect, Vardict, and Scalpel, to assess our ability to predict the allele frequencies accurately and reliably.
The results showed very high concordance between the VAFs determined by the different variant callers and what is known to be true in the reference material provided by SeraCare. We found that variations in performance do occur with different enrichment panel sizes, DNA input amounts, sequencing depth, and analysis tools. For example, while results were similar with 50 ng of input DNA and 25 ng of input DNA, enrichment panel size had a significant impact on results, with the hotspot panel having 78% specificity while the more comprehensive gene panel’s specificity was 92%. Some variability in results was also observed based on the variant calling software that was used.
Using a reference with a high number of variants enabled the determination of the average sensitivity of the assay. Additionally, the reference allowed for an interrogation of the sensitivity of the assay at challenging sites such as flanking homopolymers or at the ends of reads. This study suggests that use of well-established, highly-multiplexed reference materials during optimization and validation is critical to fine-tune assay performance.
As NEBNext Direct continues producing Custom Ready Panels ranging from single-genes to several hundred genes, the availability of well-characterized reference materials will be critical not only for the development and optimization of target-enrichment panels, but also for customers to validate and implement the panels in their own labs. As size and content will vary widely across panels, a reference with flexible content in necessary for this purpose. To respond to this need, SeraCare is now offering the VariantFlexTM system, which is a broad library of mutations to develop custom controls and reference materials for NGS-based assays. These biosynthetic constructs contain variants such as SNVs, INDELs, frameshifts, structural variants, and copy number amplifications that can be used to formulate controls across a range of VAFs, providing the necessary content and flexibility to verify assay performance.
Weren’t able to make it to AMP?
Don’t worry, we recorded a video of the poster presentation. Click here to access both the video and a PDF of the poster.