A recent study, published in the Journal of Molecular Diagnostics, describes a new, more sensitive Hepatitis B Virus (HBV) assay1. This study, led by Song-Mei Liu, MD, PhD, of the Center for Gene Diagnosis, Zhongnan Hospital of Wuhan University in China, is particularly exciting because the new assay can be used to diagnose hepatocellular carcinoma (HCC) at an earlier stage and to manage antiviral HBV treatments more effectively. It also highlights the way innovative molecular diagnostics can play a synergistic role with the development of new pharmaceutical therapeutics.
At least 240 million people are estimated to be chronically infected with HBV worldwide2. About 95% of adults who acquire HBV will spontaneously clear the infection, but for the remaining ~5%, and those who acquire the infection at birth, the virus becomes a persistent, lifelong infection3. Chronic HBV infection causes liver damage, including cirrhosis, is strongly associated with the development of hepatocellular carcinoma, and accounts for approximately 1 million deaths annually3. Although there is a vaccine to prevent infection, once a person becomes chronically infected, there is no true cure currently available.
cccDNA Allows Infection to Persist Despite Treatment
Anti-viral therapeutics can effectively clear the viral load circulating in the bloodstream, but HBV can persist and evade the immune system because there is a reservoir of the viral genome inside the nuclei of infected hepatocytes. This reservoir is the covalently closed circular DNA (cccDNA) form of the HBV genome, which is like a plasmid and exists as a stable minichromosome in the nucleus. The antiviral defense mechanisms of the immune system may not be very effective against cccDNA because it is fully histone bound4, uses host cell transcription machinery, and limits the copy number to approximately 1 -3 per cell nuclei5. Even after clearance of viral particles from the blood to non-detectable levels, treatment cessation results in relapse because of this reservoir, so a true cure of chronic HBV infection would require elimination of cccDNA from infected hepatocytes.
The life cycle of the HBV virus is shown in Figure 1 and shows the central role of cccDNA as the template for all viral RNA transcription and for progeny viruses. In circulation, the virus particles contain relaxed circular DNA (rcDNA) that is only partially double stranded. The hepatocytes in the liver are the only cells susceptible to infection, and the virus binds a hepatocyte specific receptor to gain entry. The nucleocapsids are transported to the nuclear membrane and the rcDNA is released into the nucleus. The rcDNA is converted to supercoiled cccDNA through multiple steps including removal of the viral polymerase, removal of an RNA primer from the positive strand, repair of the incomplete positive strand, and ligation of both DNA strands2. The cccDNA binds both histone and non-histone proteins and is compacted in a way that resembles cellular chromatin. Five viral RNAs encoding viral antigens are transcribed from the cccDNA , as well as a large pregenomic RNA (pgRNA). The RNAs are exported to the cytoplasm where nucleocapsid proteins are translated and assembled; reverse transcription of the pgRNA is “hidden” because it takes place within the newly formed nucleocapsids6. Many of the currently approved treatments for HBV are nucleoside analogs (NAs) that inhibit this reverse transcription step, so they may reduce the level of viremia even below detection limits, but they do not directly target destruction of the cccDNA. After second strand synthesis, the nucleocapsids with the mature relaxed circular DNA can be enveloped and exported from the cell as progeny viruses or they can be recycled back to the nucleus to replenish the cccDNA pool.
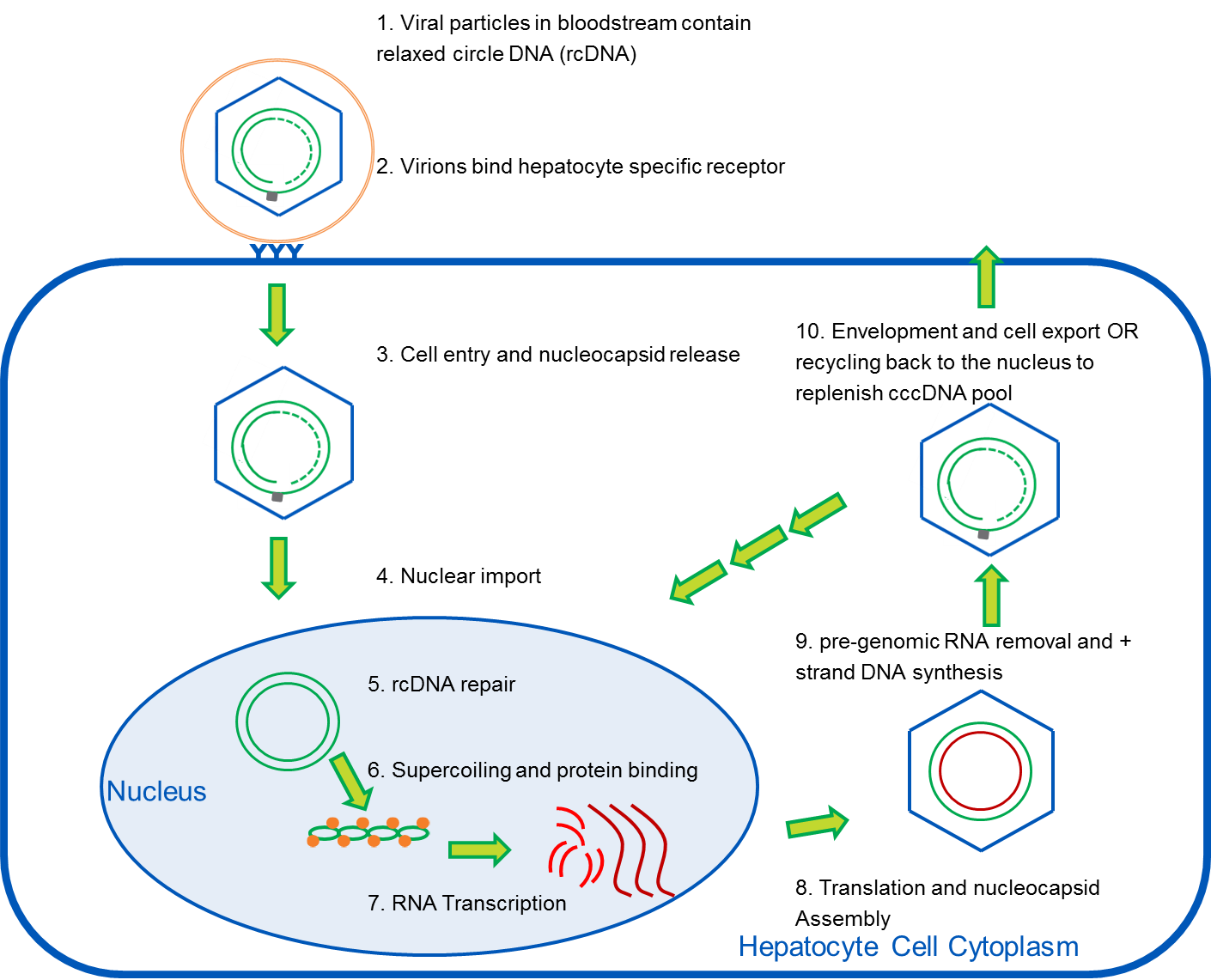
Figure 1: Life Cycle of HBV virus. Adapted from references 2 and 3.
New Therapeutics Target cccDNA Destruction
Since cccDNA is such a key intermediate in the viral life cycle, a new generation of drugs are in development to target destruction of cccDNA and effect a true cure. For example, CRISPR/cas and TALEN are genome editing systems which can be used to attack and destroy the cccDNA, but delivering them effectively to the infected viral cell nucleus remains a challenge7. Other new therapeutics are focusing on ways to upregulate APOBEC3A and APOBEC3B cytidine deaminases in the infected nuclei. Such activation could generate an apurinic/apyrimidinic site - a DNA gap that would trigger cccDNA degradation8. A potential vulnerability of the cccDNA minichromosome is that is does not contain a centromere or a nuclear retention signal so can be excluded from the nucleus after mitosis and lost as cells divide. Stimulation of the immune, including CD8+ T cells and natural killer cells, can kill infected cells and destroy the cccDNA inside, but can also induce proliferation of the neighboring hepatocytes to compensate for the cell loss, and the cell division may further reduce the pool of cccDNA2. Cytokines that reduce the recycling of progeny viruses back to the nucleus can further deplete the cccDNA pool. So, combination therapy that acts through both cytolytic and non-cytolytic cytokine-mediated mechanisms appears promising for cccDNA clearance.
Sensitive Quantitation of cccDNA Is Needed
Since measuring the pool of cccDNA is so important for therapeutic outcomes, new diagnostic tests specific for the cccDNA are needed. A major step forward was the development of PCR based tests that discriminate cccDNA from linear viral DNA or relaxed circle DNA that are present in vast excess. These PCR methods have primers and/or probes that span the nick in rcDNA and hybridize to the “gap region” that is not double stranded. Takkenberg and colleagues9 developed a real-time PCR assay for detection and quantitation of cccDNA in plasma and validated their assay using a 15-member HBV genotype panel from SeraCare. This panel contains undiluted plasma from patients with high viral load collected worldwide across genotypes A-H. Detection (and quantitation) of cccDNA did not appear to correlate with HBV genotype; however, assay sensitivity was not optimal and cccDNA could not be detected in ~43% of the samples tested.
Now, detection methods have been taken one step further in the publication “A Highly Sensitive and Robust Method for Hepatitis B Virus Covalently Closed Circular DNA Detection in Single Cells and Serum” published in the April 2018 issue of Journal of Molecular Diagnostics1. The group developed a droplet digital PCR (ddPCR) based method which is more sensitive for detecting trace molecules (more effective at finding the needle in a haystack). They used cccDNA selective primers and increased the sensitivity and accuracy by using Plasmid-Safe™ ATP-Dependent DNase to eliminate rcDNA, single-stranded DNA, and double-stranded linear DNA. The authors collected serum from 79 non-HCC patients and 89 patients with HCC. In addition to testing for cccDNA, data on viral load, serological HBsAg levels and antibody levels were collected. Serum cccDNA copy number was higher in HCC patients compared to non-HCC patients. The more sensitive quantification of serum cccDNA along with HBV-DNA viral load was effective in discriminating HCC patients from non-HCC patients, with areas under the curve of receiver operating characteristic of 0.847 and an assay sensitivity of 74.5% and specificity of 93.7%. This finding holds promise for diagnosing HCC at an earlier stage without the need for liver biopsy. Another advantage of a liquid biopsy approach is that the number of cccDNA molecules can vary significantly from one infected hepatocyte to the next within the same patient, but serum and plasma have a homogenous cccDNA distribution
Panels and Reference Materials Can Help
As Fan Shen mentions in the commentary that accompanies the JMD publication10, “Genetic diagnostics is becoming a conventional practice in the field of hepatology” and although ddPCR is entering numerous laboratories worldwide, the clinical translation is still pending. Large panels of characterized HBV positive serum and plasma can be invaluable in the clinical validation of new molecular diagnostic tests. SeraCare offers AccuSet™ HBV Worldwide Performance Panel and AccuSet HBsAg Performance Panel which are 7-member and 25-member panels of undiluted, naturally occurring plasma samples. Each panel contains a comprehensive comparative data sheet with test results from a wide variety of assays and platforms, and demographic information is also available. These performance panels can be used to evaluate assay specificity, sensitivity, repeatability, and reproducibility to assist in validating new test methods and equipment.
For a limited time, get 20% off all SeraCare panels. Offer good until September 30, 2018.
References
- Huang JT, Yang Y, Hu YM, Liu XH, Liao MY, Morgan R, Yuan EF, Li X, Liu SM. A Highly Sensitive and Robust Method for Hepatitis B Virus Covalently Closed Circular DNA Detection in Single Cells and Serum. J Mol Diagn. 2018 May;20(3):334-343.
- Allweiss L, Dandri M. The Role of cccDNA in HBV Maintenance. Viruses. 2017 Jun 21;9(6).
- Grimm D, Thimme R, Blum HE. HBV life cycle and novel drug targets. Hepatol Int. 2011 Jun;5(2):644-53.
- Newbold JE, Xin H, Tencza M, Sherman G, Dean J, Bowden S, Locarnini S. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J Virol. 1995 Jun;69(6):3350-7.
- Laras A, Koskinas J, Dimou E, Kostamena A, Hadziyannis SJ. Intrahepatic levels and replicative activity of covalently closed circular hepatitis B virus DNA in chronically infected patients. 2006 Sep;44(3):694-702.
- Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008 Jun;134(1-2):235-49.
- Lucifora J, Protzer U. Attacking hepatitis B virus cccDNA--The holy grail to hepatitis B cure. J Hepatol. 2016 Apr;64(1 Suppl):S41-S48.
- Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Hüser N, Durantel D, Liang TJ, Münk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. 2014 Mar 14;343(6176):1221-8.
- Takkenberg RB, Menting S, Beld MG. Validation of a sensitive and specific real-time PCR for detection and quantitation of hepatitis B virus covalently closed circular DNA in plasma of chronic hepatitis B patients. Methods Mol Biol. 2012;903:113-28.
- Shen F, Sergi C, Sun HL. Hepatitis B Virus Covalently Closed Circular DNA-Selective Droplet Digital PCR: A Sensitive and Noninvasive Method for Hepatocellular Carcinoma Diagnosis? J Mol Diagn. 2018 May;20(3):277-278.





