Choose your Article Focus | NGS | Molecular & Serology
A Multi-Lab Study of Fusion RNA Reference Standards Using Amplicon- and Hybrid Capture-based Targeted NGS Panels
Category: NGS, Assay Development, RNA fusion
Posted by
Andrew Anfora, PhD on Jan 28, 2020 12:00:00 AM
Introduction Sourcing assay validation samples as positive run controls or workflow controls in targeted NGS RNA fusion assays remains a challenge today. This is further exacerbated with clinical labs looking to provide validated NGS assays for patient stratification in a host of new drugs in clinical trials or newly approved targeting fusion genes, such as NTRK genes (Larotrectinib, Loxo/Bayer) and Entrectinib (Genetech/Roche) for rare cancers in adult and pediatric patients, and RET (Loxo/Lilly) for lung cancer. SeraCare produces several RNA fusion reference materials. This article describes the development and multi-laboratory evaluation of a pan-cancer multiplexed Fusion RNA reference standard for analysis of clinically relevant fusion genes in solid tumors. The evaluation was conducted at 5 different laboratories on different NGS platforms (amplicon- and hybridization capture-based) as well as at different RNA inputs within a platform. Results highlight the utility of this Fusion RNA reference material to support clinical NGS assays as positive controls in solid tumor cancer patient stratification for many of these fusion-based targeted therapies. Data Discussion Fusion RNA Reference Standard Development 18 clinically actionable biosynthetic fusion genes were transcribed and blended with reference cell line human cell line (GM24385, from GIAB), The RNA fusion constructs in the Fusion RNA reference material were blended at ~1500 copies per uL, and measured by digital droplet PCR (ddPCR) measurements as well as by an orthogonal targeted NGS assay (Archer FusionPlex Solid Tumor and MiSeq v2 300 cycle kit). Results are shown in Figure 1 below. Figure 1: Concentration (blue) and number of unique start sites (green) measured for each fusion RNA in the fusion reference material as determined by RT-dPCR and targeted NGS panel, respectively. Data represent average of three replicate measurements. As shown in Figure 1, all 18 gene fusions in the Fusion RNA reference material were detected on the Archer ST assay, at different levels of abundance. Approximately 50% of the variants showed similar relative abundance between ddPCR and NGS measurements. ~25% of the variants showed an apparent increase in relative abundance as measured by ddPCR, and the remaining ~25% of the gene fusions showed an increase in relative abundance as measured by NGS. A multi-laboratory evaluation of the Fusion RNA reference material was conducted at 5 laboratories (Site A – E, see Table 1). However, this article will focus on evaluations involving the Archer FusionPlex Solid Tumor and custom Solid Tumor-based targeted NGS panels. For discussion of all data generated by all laboratories listed in Table 1, check out our poster presentation from AACR 2018. Table 1: Assay, input, and analysis used at Site A – E. Figure 2 demonstrates the reproducibility of the Archer FusionPlex Solid Tumor assay across multiple sites and operators from the highly multiplexed contrived Fusion RNA reference material. The discordant data points are likely due to a combination of different operators, use of different analysis software versions, and differences in replicate measurements between sites. To account for the multitude of panel-based assays, one laboratory used a custom panel assay to evaluate the RNA fusion reference material. Beta test Site D used the fusion reference material with a customized Archer FusionPlex assay as shown in Figure 3. The functionality of the customized panel was clearly demonstrated, in addition, a limit of detection study was conducted with the abundant reference material, with most fusions detected at all tested input amounts.[1] [1] For another RNA fusion limit of detection study see our AACR 2019 poster. Summary SeraCare has developed a reference material that contains 18 clinically relevant RNA fusions. The RNA fusion reference material was evaluated at 6 different laboratories. Results of these analysis highlight the robustness of the Seraseq Fusion RNA reference materials in multi-lab concordance studies as well as a tool uniquely suited for limit of detection (LoD) studies. The RNA fusion reference material is also used in analytical validation of targeted NGS panels, and as an objective reference standard for inter-laboratory comparison of NGS assay performance. A similar multi-lab evaluation of the FFPE Fusion RNA reference materials will be published shortly.
0 Comments Click here to read/write comments
Developing a Rock-Solid Lung Cancer Assay, Part 2
Category: NGS, cancer, Assay Development, Lung Cancer
Posted by
Yves Konigshofer, PhD on Jul 2, 2018 12:00:00 AM
In a recent post, we discussed key considerations for designing a robust next-generation sequencing (NGS)-based lung cancer assay. Putting those plans into action in the development phase brings forth a new set of challenges. Through our experience developing NGS reference materials and the relationships we’ve built with assay developers of all stripes, we’ve identified those important factors and ways to navigate them. But before you begin designing and optimizing your assay, you should become very familiar with binomial and Poisson distributions and their use because the outcome of many analytical steps can be modeled and explained with them.
0 Comments Click here to read/write comments
How am I going to test my assay? Should I use patient samples or biosynthetic materials?
Category: NGS, Assay Development
Posted by
Dan Brudzewsky on Jan 4, 2018 12:00:00 AM
Assay development and optimization for clinical genetics is increasingly challenging. In an era of clinical genomics, new technologies and clinical utilities constantly call for newer and better performing assays. Having access to an abundant supply of relevant and reliable test material is critical for quick assay development and well-documented assay performance.
0 Comments Click here to read/write comments
Accelerating Liquid Biopsy Assay Development
Category: liquid biopsy, Assay Development, ctDNA, reference materials
Posted by
Sam Blier on Sep 8, 2017 12:00:00 AM
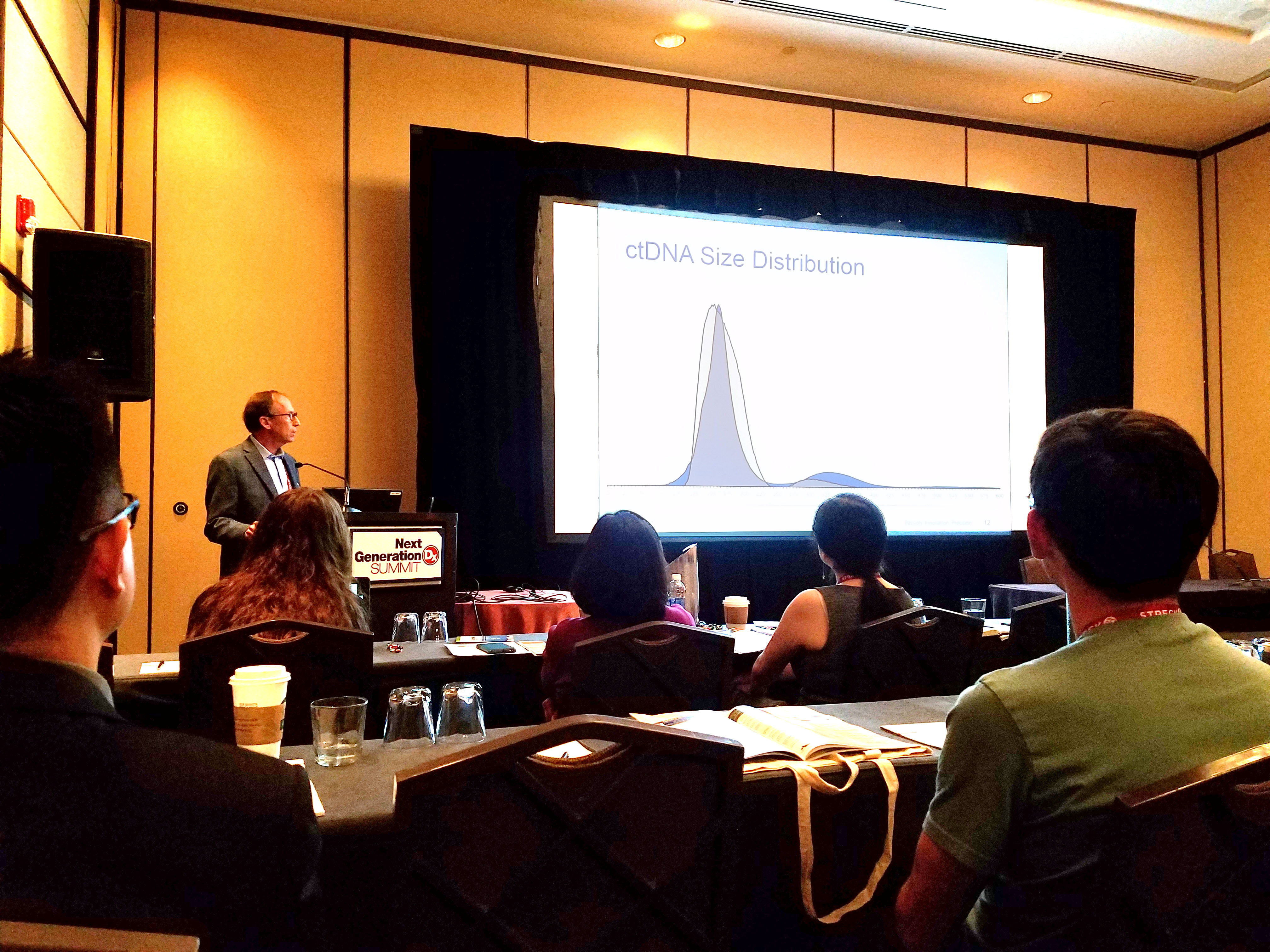
At the 2017 Next Generation Dx Summit in Washington, DC, our CSO, Russell Garlick, PhD, presented a workshop on accelerating liquid biopsy assay development. He has worked closely with a variety of groups in the liquid biopsy space that are developing and validating circulating tumor DNA (ctDNA) assays. He highlighted some common challenges facing the field, and explained how SeraCare has been using these collaborations to develop QC tools specifically for ensuring the robustness of these cutting-edge tests.
0 Comments Click here to read/write comments
How Many Target Copies Are Present in Your Plasma DNA Sample?
Category: liquid biopsy, cancer, Assay Development, ctDNA
Posted by
Dale Yuzuki on Jun 15, 2017 12:00:00 AM
The field of circulating tumor DNA analysis (ctDNA, also sometimes called in a larger context “liquid biopsy”) holds great promise for monitoring response to cancer treatment, assisting therapeutic choice, monitoring recurrence, and for pre-cancer screening. As such there is a great amount of assay development and ongoing clinical trials; at ClinicalTrials.gov searching for the term "Circulating DNA" you can find over 180 open clinical trials for a wide range of tumor types and interventions.
0 Comments Click here to read/write comments