The field of circulating tumor DNA analysis (ctDNA, also sometimes called in a larger context “liquid biopsy”) holds great promise for monitoring response to cancer treatment, assisting therapeutic choice, monitoring recurrence, and for pre-cancer screening. As such there is a great amount of assay development and ongoing clinical trials; at ClinicalTrials.gov searching for the term "Circulating DNA" you can find over 180 open clinical trials for a wide range of tumor types and interventions.
Yet experts in the field indicate that the median allele frequency of ctDNA of cancer patients is less than 1% of the total circulating cell free DNA (ccfDNA) in the blood. How much ctDNA can you derive from a person who is suffering from a deadly disease?
Healthy plasma typically yields 5 to 10 ng of ccfDNA per mL of plasma when properly collected and processed. However in cancer patients, the amount of detectable ctDNA varies widely. Table 1 summarizes data from an influential 2014 study from Johns Hopkins that looked at 640 patients with various cancer types using a sensitive digital PCR method called BEAMing, and the fraction of patients with detectable DNA.
Table 1: Data from Bettegowda et al Sci Trans Med 2014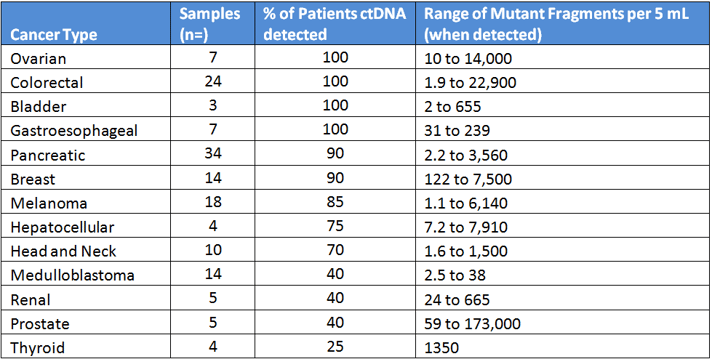
The amounts of ctDNA from these patients were highly variable, even with those of the same tumor type. This underscores the need for high sensitivity of these assays.
Combining these ideas, if a cancer patient has on the order of 5 ng of ccfDNA (not high molecular weight genomic DNA) in a milliliter of plasma, there are only 3,000 genome equivalents present. On top of this, a high proportion of the ctDNA is fragmented into periodic nucleosomal wrap units of about 170 bp and a minor peak at about 340 bp; another challenge is that given the nature of the fragmented DNA, a PCR amplicon or hybridization-capture probe may not have sufficient overlap to capture a given short molecule.
And in addition, consider the problem of NGS assay efficiency throughout the entire process; focused digital PCR assays will have much higher detection sensitivity, down to a single molecule detection, at the expense of limited numbers of mutations tested for simultaneously.
Potential losses of sensitivity come from many sources in an NGS-based assay: from extraction, from PCR efficiency (i.e. what percentage of molecules available for amplification actually do amplify), to library construction with its attendant manipulations and clean-up steps (with multiplicative losses along the way), to cluster or ISP bead generation (a minor loss), to sequence generation. With a given number of molecules at the input level, the efficiency of target molecules going through this process could be 80% or 50%; what is measured at the end is only a ratio of mutant to wild-type, characterized by allele frequency. It is not a measure of absolute quantitation.
The number of total available molecules for detection is moot where the input amounts of DNA are relatively abundant and the desired mutant molecules are also present in sufficient numbers. However in the case of ctDNA, the amounts of total available ccfDNA may vary widely, and if the mutant DNA is a very small percentage of the total available these considerations take on much greater significance.
In a recent SeraCare and GenomeWeb-produced webinar, Dr. Tony Godfrey (Boston University School of Medicine, Boston MA) illustrated this point with a chart with calculation of input DNA (in nanograms) versus the total copies and number of mutant copies down to 0.01%, reproduced and expanded below in Table 2.
Table 2: Total and Mutant Copies available as a function of input DNA
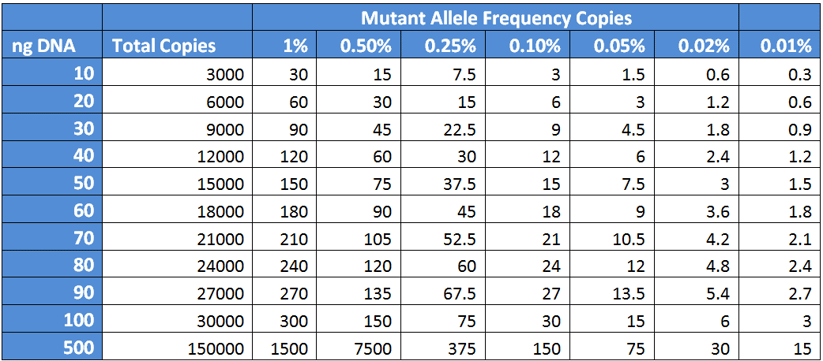
What you can readily observe is at 0.1% mutant allele frequency, you will need plenty of input DNA (on the order of 30 to 50 ng) in order to get 9 to 15 copies of target. To reiterate, the fragmented nature of ctDNA will mean these 9 to 15 target copies may not all be intact, and even with an intact target for the PCR or hybridization-capture method of enrichment there will be additional detection losses.
SeraCare carries a range of ctDNA reference materials at multiple allele frequencies to enable accurate assay development and optimization, and to enable assay validation. In addition to a purified DNA format, SeraCare has a stabilized and patient-like ctDNA reference material to assess the performance of your sample-to-answer laboratory process. For more information click here.





