Choose your Article Focus | NGS | Molecular & Serology
Assessing RNA Extraction with FFPE Fusion RNA Reference Materials
Category: NGS, RNA fusion, reference materials
Posted by
Dan Brudzewsky on Mar 11, 2020 12:00:00 AM
This is a third blog in a series on RNA fusions, this time focusing on how the FFPE Fusion RNA materials are used as RNA extraction controls.
0 Comments Click here to read/write comments
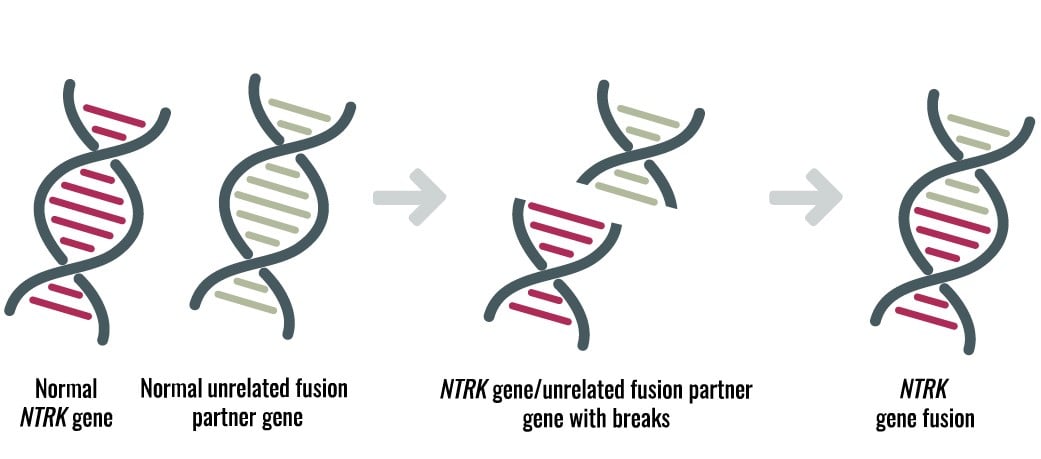
Diagnostic Testing Schemes for NTRK Cancers: All Roads Lead to NGS
Category: SeraSeq, NTRK, NGS, RNA fusion
Posted by
Russell Garlick, PhD on Feb 12, 2020 12:00:00 AM
New treatment options for cancer patients with neurotrophic tyrosine kinase (NTRK) rearrangements are a tremendous success, demonstrating first-hand the importance of precision diagnostics. The FDA granted accelerated approval of Bayer's VITRAKVI (larotrectinib) for adult and pediatric solid tumors containing NTRK fusions1. The approval was one of the first tissue agnostic approvals based on genotyping results, and included patients with unresectable or metastatic cancer. For 12 cancer types, the overall response rate was 75% with 22% complete response and 53% partial response. In addition to VITRAKVI, Genentech's entrectinib has a "breakthrough" status and is being evaluated for the potential treatment of advanced or metastatic tumors that harbor NTRK or ROS1 gene rearrangements.
0 Comments Click here to read/write comments
Multi-Lab Study of Fusion RNA Reference Standards for Targeted NGS
Category: NTRK, NGS, RNA fusion, reference materials, AACR
Posted by
Andrew Anfora, PhD on Jan 28, 2020 12:00:00 AM
Sourcing assay validation samples as positive run controls or workflow controls in targeted NGS RNA fusion assays remains a challenge today. This is further exacerbated with clinical labs looking to provide validated NGS assays for patient stratification in a host of new drugs in clinical trials or newly approved targeting fusion genes, such as NTRK genes (Larotrectinib, Loxo/Bayer) and Entrectinib (Genentech/Roche) for rare cancers in adult and pediatric patients, and RET (Loxo/Lilly) for lung cancer. SeraCare produces several RNA fusion reference materials. This article describes the development and multi-laboratory evaluation of a pan-cancer multiplexed Fusion RNA reference standard for analysis of clinically relevant fusion genes in solid tumors. The evaluation was conducted at 5 different laboratories on different NGS platforms (amplicon- and hybridization capture-based) as well as at different RNA inputs within a platform. Results highlight the utility of this Fusion RNA reference material to support clinical NGS assays as positive controls in solid tumor cancer patient stratification for many of these fusion-based targeted therapies.
0 Comments Click here to read/write comments
A Multi-Lab Study of Fusion RNA Reference Standards Using Amplicon- and Hybrid Capture-based Targeted NGS Panels
Category: NGS, Assay Development, RNA fusion
Posted by
Andrew Anfora, PhD on Jan 28, 2020 12:00:00 AM
Introduction Sourcing assay validation samples as positive run controls or workflow controls in targeted NGS RNA fusion assays remains a challenge today. This is further exacerbated with clinical labs looking to provide validated NGS assays for patient stratification in a host of new drugs in clinical trials or newly approved targeting fusion genes, such as NTRK genes (Larotrectinib, Loxo/Bayer) and Entrectinib (Genetech/Roche) for rare cancers in adult and pediatric patients, and RET (Loxo/Lilly) for lung cancer. SeraCare produces several RNA fusion reference materials. This article describes the development and multi-laboratory evaluation of a pan-cancer multiplexed Fusion RNA reference standard for analysis of clinically relevant fusion genes in solid tumors. The evaluation was conducted at 5 different laboratories on different NGS platforms (amplicon- and hybridization capture-based) as well as at different RNA inputs within a platform. Results highlight the utility of this Fusion RNA reference material to support clinical NGS assays as positive controls in solid tumor cancer patient stratification for many of these fusion-based targeted therapies. Data Discussion Fusion RNA Reference Standard Development 18 clinically actionable biosynthetic fusion genes were transcribed and blended with reference cell line human cell line (GM24385, from GIAB), The RNA fusion constructs in the Fusion RNA reference material were blended at ~1500 copies per uL, and measured by digital droplet PCR (ddPCR) measurements as well as by an orthogonal targeted NGS assay (Archer FusionPlex Solid Tumor and MiSeq v2 300 cycle kit). Results are shown in Figure 1 below. Figure 1: Concentration (blue) and number of unique start sites (green) measured for each fusion RNA in the fusion reference material as determined by RT-dPCR and targeted NGS panel, respectively. Data represent average of three replicate measurements. As shown in Figure 1, all 18 gene fusions in the Fusion RNA reference material were detected on the Archer ST assay, at different levels of abundance. Approximately 50% of the variants showed similar relative abundance between ddPCR and NGS measurements. ~25% of the variants showed an apparent increase in relative abundance as measured by ddPCR, and the remaining ~25% of the gene fusions showed an increase in relative abundance as measured by NGS. A multi-laboratory evaluation of the Fusion RNA reference material was conducted at 5 laboratories (Site A – E, see Table 1). However, this article will focus on evaluations involving the Archer FusionPlex Solid Tumor and custom Solid Tumor-based targeted NGS panels. For discussion of all data generated by all laboratories listed in Table 1, check out our poster presentation from AACR 2018. Table 1: Assay, input, and analysis used at Site A – E. Figure 2 demonstrates the reproducibility of the Archer FusionPlex Solid Tumor assay across multiple sites and operators from the highly multiplexed contrived Fusion RNA reference material. The discordant data points are likely due to a combination of different operators, use of different analysis software versions, and differences in replicate measurements between sites. To account for the multitude of panel-based assays, one laboratory used a custom panel assay to evaluate the RNA fusion reference material. Beta test Site D used the fusion reference material with a customized Archer FusionPlex assay as shown in Figure 3. The functionality of the customized panel was clearly demonstrated, in addition, a limit of detection study was conducted with the abundant reference material, with most fusions detected at all tested input amounts.[1] [1] For another RNA fusion limit of detection study see our AACR 2019 poster. Summary SeraCare has developed a reference material that contains 18 clinically relevant RNA fusions. The RNA fusion reference material was evaluated at 6 different laboratories. Results of these analysis highlight the robustness of the Seraseq Fusion RNA reference materials in multi-lab concordance studies as well as a tool uniquely suited for limit of detection (LoD) studies. The RNA fusion reference material is also used in analytical validation of targeted NGS panels, and as an objective reference standard for inter-laboratory comparison of NGS assay performance. A similar multi-lab evaluation of the FFPE Fusion RNA reference materials will be published shortly.
0 Comments Click here to read/write comments
So Many Posters, So Little Time
Category: TMB, RNA fusion, ctDNA, AACR
Posted by
Sam Blier on Jun 6, 2019 12:00:00 AM
Cancer research is purposely methodical and measured. So – somewhat paradoxically – it can be difficult to keep up with the steady stream of discoveries in the literature and presented at conferences like AACR. As a developer and manufacturer of platform-agnostic NGS reference standards, we’re in a unique position to collaborate with cancer genomics assay developers, laboratories, pharmaceutical companies, and other organizations invested in more precise and robust cancer tests.
0 Comments Click here to read/write comments
Presenting NTRK Reference Materials for Global Assay Standardization at AACR 2019
Category: SeraSeq, NTRK, RNA fusion, AACR
Posted by
Catherine Huang, PhD on Apr 8, 2019 12:00:00 AM
On the last morning of AACR 2019, I had the privilege of presenting a poster together with my colleague, Sebastian Bender from Bayer AG, in Berlin. Because of this, I didn’t have a chance to attend any talks, but I still wanted to finish out my blog series with highlights from each day of the conference.
0 Comments Click here to read/write comments
NTRK Fusion Testing for New Therapies
Category: NTRK, cancer, RNA fusion
Posted by
Omo Clement, PhD on Feb 6, 2019 12:00:00 AM
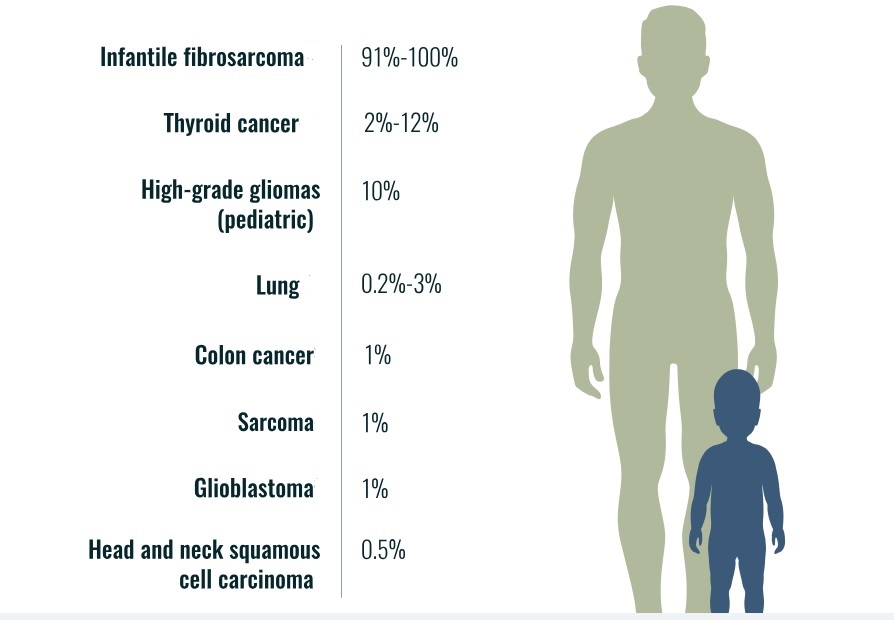
Neurotrophic tyrosine receptor kinases (NTRK) can become abnormally fused to other genes resulting in growth signals that can lead to cancer in many organs of the human body. TRK gene fusion-based cancers are rare but present in pediatric and adult cancers such as lung, thyroid, colon, etc. (see, e.g., Figure 1).
0 Comments Click here to read/write comments
Twists and Turns That Lead From “Curiosity Driven Research” To Innovative Diagnostics
Category: RNA fusion, AACR
Posted by
Catherine Huang, PhD on May 8, 2018 12:00:00 AM
Jennifer Doudna is not a cancer biologist and joked that she might deliver her entire lecture at the 2018 AACR Annual Meeting without ever mentioning the word “cancer.” However, when presenting the Irving Weinstein Foundation Distinguished Lecture, she told a fascinating story about how curiosity regarding an interesting sequence motif in bacteria led to gene-editing tools, and how investigation of the mechanisms behind those tools may lead to innovative diagnostics for the future.
0 Comments Click here to read/write comments
With so many options, how do you select the best NGS cancer assay?
Category: NGS, cancer, RNA fusion, reference materials
Posted by
Catherine Huang, PhD on Jan 11, 2018 12:00:00 AM
Clinical labs must constantly evolve their test offerings in order to support the most recent advances in clinical care. For next-generation sequencing (NGS) tumor profiling assays, there are often multiple commercially available kits with similar claims for gene content and sensitivity, as well as customized solutions. How can you quickly perform an effective evaluation of available assay systems to make a data-driven choice?
0 Comments Click here to read/write comments
3 Steps for Building a Bulletproof Clinical NGS Assay: Step 3
Category: qc management, NGS, RNA fusion
Posted by
Russell Garlick, PhD on Dec 8, 2017 12:00:00 AM
What does it mean for an NGS assay to be bulletproof and why does your lab need it? In two previous blog articles (parts one and two), we’ve talked about the factors that go into making NGS assays that doctors can rely on to deliver targeted, lifesaving therapies to their patients. Bulletproof assays are the tests that make your lab a trusted name in the NGS field, a leader in a rapidly-growing market. But, as we’ve written, genetic sequencing is complex, expensive, and time-consuming. Therefore, finding ways to do it more efficiently, while maintaining the quality of your tests, is in the best interests of your lab and its customers. As a refresher, here are the three steps for building a bulletproof clinical NGS assay: Consulting with experts Outlining your validation and quality control (QC) strategies together Evaluating reference material options We’ve already covered the first two steps. In this article, we’ll look at the third one. Choosing the right reference material technology can help control the high validation and running costs of highly multiplexed assays.
0 Comments Click here to read/write comments