On the last morning of AACR 2019, I had the privilege of presenting a poster together with my colleague, Sebastian Bender from Bayer AG, in Berlin. Because of this, I didn’t have a chance to attend any talks, but I still wanted to finish out my blog series with highlights from each day of the conference. The poster session was a personal highlight of the meeting because of all the great feedback we received on the project. I want to share with you a little about the poster we presented called “Development of NTRK Reference Materials for Global Assay Standardization,” which is available for you to download along with our other featured posters.
This poster came out of a collaboration between SeraCare, Bayer AG, and Bayer US that identification of genomic alterations underlying carcinogenesis through genomic testing is critical to match patients to either an approved cancer therapy or a drug in a clinical trial. Chromosomal rearrangements of the three neurotrophic tyrosine receptor kinase genes (NTRK1, NTRK2, NTRK3) that encode tropomyosin receptor kinases (TRK) are genomic alterations that drive oncogenesis in many adult and pediatric cancers. In TRK fusion cancer, the NTRK gene fuses with an unrelated gene, causing overexpression of the oncogenic TRK fusion protein.
Image for illustrative purposes only. Courtesy of www.trkcancer.com.
Accurate and reliable genomic testing for NTRK fusions has become critically important because of innovative precision biopharmaceutical treatments such as larotrectinib, which was approved by the US Food and Drug Administration (FDA) for the treatment of patients with locally advanced or metastatic solid tumors harboring NTRK gene fusions. In this study, we aimed to develop a 15-plex reference standard which can be used to evaluate genomic NTRK testing, optimize assays, tune bioinformatics pipelines, and benchmark among testing sites.
The criteria for selecting the 15 NTRK fusion RNAs in the reference standard included:
- Fusions involving all three NTRK genes (five fusions each for NTRK1, NTRK2, and NTRK3).
- Prevalence data for NTRK fusions published in three larotrectinib studies (Drilon et al. 2018; NEJM; Table S2) with all selected fusions present in at least two patients.
- Prevalence data for NTRK fusions in the COSMIC database (as of May 2018).
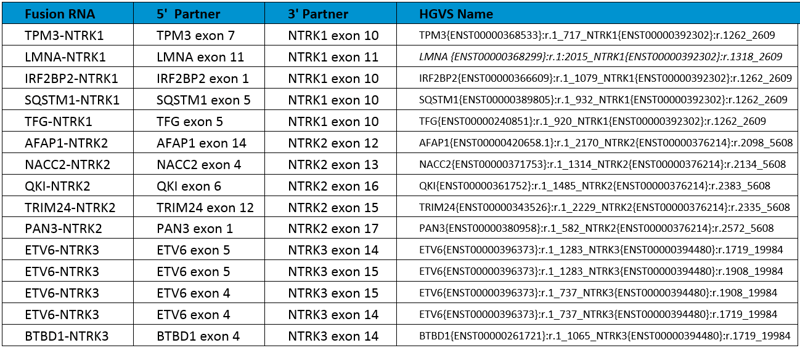
Overview of all 15 NTRK fusion RNAs included in the reference standard.
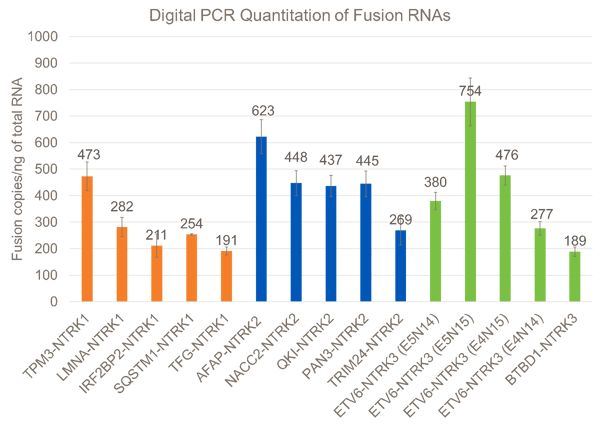
After the NTRK fusions were selected, DNA constructs containing the fusions of interest were synthesized, and biosynthetic fusion RNAs were transcribed in vitro. GM24385 cell line was modified to contain the biosynthetic NTRK RNA fusions. Cells were fixed in 10% buffered formalin for 18 hours, embedded in paraffin, and 10-micron curls were sectioned. Curls were extracted and tested using in-house developed digital PCR assays run on Bio-Rad Qx200™. Digital PCR is an absolute quantitation method and gives an accurate measure of the copies of each biosynthetic fusion RNA per nanogram of total RNA extractable from the curl.
Archer® FusionPlex® Solid Tumor Kit run on Illumina MiSeq® and OncomineTM Comprehensive Assay were used to demonstrate that the reference standard is compatible with commonly used commercial NGS assays. You can download the poster here to review the results.
There was a lot of discussion among poster attendees about why the percent of reads supporting the fusions varies so much on the Archer assay (6.2% for LMNA-NTRK1 up to 63.1% for AFAP1-NTRK2). The Archer analysis software defines this metric as “percent of reads supporting the event. It is the number of unique reads spanning the breakpoints and supporting the event, divided by the total number of unique RNA reads that span either breakpoint. Reads from all primers are combined for this calculation (but only those reads spanning the breakpoint on either side of the junction).” So, it appears that if both of the fusion partner transcripts are poorly expressed in the GM24385 reference cell lines, most of the reads will be from the biosynthetic fusion transcripts, and the percent of reads supporting the fusion will be high. However, if either of the partner transcripts are highly expressed in GM24385 (like LMNA is), then many reads will come from the endogenous transcripts and the percent of reads supporting the fusion will be low.
Another thing I learned from conversations at this poster is that labs are using a wide variety of assays beyond the Archer and Oncomine platforms -- some of which score for 5’ and 3’ imbalance of exon expression across key transcripts. I gained so much insight and knowledge in Atlanta, and I’m hoping to gather some more data for this product on diverse testing systems in our lab.
The key takeaways from our poster were that high-quality genomic cancer testing is essential for identifying targetable alterations in cancer patients who may benefit from precision therapies. Highly-characterized, patient-like reference standards such as the Seraseq® FFPE NTRK Fusion RNA Reference Material described here can be used to standardize testing across laboratories and clinical sites. This reference standard contains 15 clinically relevant NTRK fusions in a single reference sample and is compatible with different NGS testing methodologies.
This was one of several great posters that we and our collaborators presented at AACR on topics from TMB to liquid biopsy. You can download them all for free right here.





