At the 2017 Next Generation Dx Summit in Washington, DC, our CSO, Russell Garlick, PhD, presented a workshop on accelerating liquid biopsy assay development. He has worked closely with a variety of groups in the liquid biopsy space that are developing and validating circulating tumor DNA (ctDNA) assays. He highlighted some common challenges facing the field, and explained how SeraCare has been using these collaborations to develop QC tools specifically for ensuring the robustness of these cutting-edge tests.
Validate early and often
Dr. Garlick detailed an interesting study in which a customer was validating a liquid biopsy assay using a custom ctDNA reference material that SeraCare produced for them. Their assay was designed to detect TP53 COSM45437 c.784_786delGGT (deletion), but it was generating alternative interpretations of the same mutant allele. The SeraCare ctDNA reference material featuring their specific deletion target revealed that the amplicon was designed around one position, but the process was excluding the second position because it was on the edge of the amplicon. They were expecting their assay to be able to detect this, but the reference material exposed the deficiency. Since they were still in development, they were able to redesign the amplicon to ensure the assay’s robustness. By incorporating biosynthetic NGS reference materials into their development and validation process, they were able to quickly troubleshoot and solve the problem to get a more robust assay to market faster.
Ensuring clinical utility
Analyzing EGFR T790M c.2369C>T mutations in NSCLC patients is important for determining therapeutic options. But in a recent study involving four laboratories, T790M positive predictive values in plasma ranged from 20-70%. Some studies have reported T790M false-positive rates of up to 30%. This means that there are commercialized assays that aren’t fitting the needs of the clinical oncologist. Dr. Garlick pointed out that this is an ideal use-case for biosynthetic NGS reference materials. He reviewed digital PCR and NGS data from the Seraseq™ ctDNA Reference Material v2 that suggests that this observed discordance can result from spontaneous cytosine deamination. By using biosynthetic NGS reference materials for a performance study, you have a tool “you can use as a fallback position, as a stake in the ground, a truth-set.” Figures 1a and 1b show results from a droplet digital PCR assay for EGFR T790M in the Seraseq ctDNA v2 Reference Material.
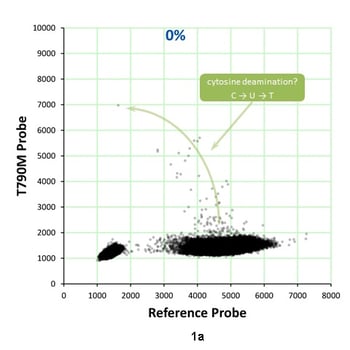
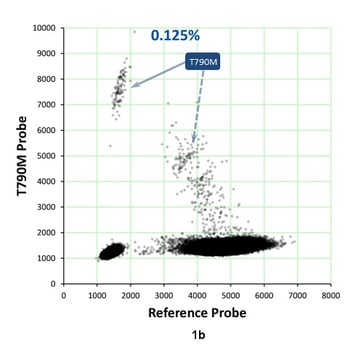
Figure 1a represents the WT GM24385 mix used to generate the 0.125% allele frequency of the T790M shown in Figure 1b. In the WT sample, there is only one droplet (in ~200,000) that is in the region of T790M single=positive droplets. With ~55% of droplets being positive for the reference sequence, only ~1 droplet would be expected that is positive for both reference and T790M. However, significantly more than one droplet appears to be double-positive. For double-positive droplets in the 0% sample, the T790M signal appears to be higher when the reference is lower, suggesting that it may form in some droplets during the first few cycles. Overall, this suggests that discordance for C>T can result from spontaneous cytosine deamination. These types of modifications can be precisely assessed using biosynthetic reference materials.
The T790M example highlights the need for tools that allow assay developers to “start testing lab-to-lab, and platform-to-platform capabilities.” Biosynthetic NGS reference materials “provide us improved and standardized measurements and will lead to better discrimination in diagnosis,” which is critical as liquid biopsies see wider clinical adoption.
Better tools for better assays
Liquid biopsy assay developers have a few options for reference materials, but Dr. Garlick shed some light on what’s driving the need for patient-like biosynthetic NGS reference materials. Circulating tumor DNA is significantly different from that found in solid tissue, so ctDNA reference materials need to be different too. Circulating cell-free DNA (cfDNA) is bound to nucleosome complexes, and averages around 166 bp, which means that liquid biopsy assays are designed to detect minute amounts of specifically sized ctDNA surrounded by lipophilic materials from the apoptotic fractions. Without a reference material that accurately represents the native cfDNA being analyzed, assay performance can be compromised. Some developers have turned to Genome-In-A-Bottle type reference materials, but a recent JMD article pointed out that you cannot use GIAB-type reference materials for insertions/deletions (INDELs), which limits the feasibility of that approach. In addition, input mass into any liquid biopsy assay is critical. A recent study showed greater than 80% extraction (Figure 2) efficiency of the Seraseq ctDNA Reference Material v2, which is comparable to that of patient specimens. So these reference materials provide an excellent way to evaluate how input mass affects your assay’s performance by providing a direct correlation to how you can expect your assay to perform in the field.
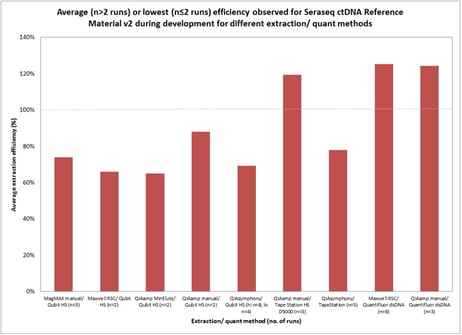
Figure 2: These data are from the extraction study that used prototype ctDNA v2 reference material with 40 variants present at 0.18% allele frequency. Note that yield appears to be >100% for several of the extraction/quant combinations – this is expected given the rather arbitrary designation of the reference methods.
For an in-depth look at how these materials are made, including performance data, download this free tech note.





