Previously, we wrote about some of the Quality Control challenges that clinical laboratories performing Next Generation Sequencing (NGS) face towards ensuring their assays are safe and effective for guiding medical management decisions. Reliable access to high quality reference materials is necessary to help overcome these challenges; however, it is not sufficient. Insights that reference materials provide into the health of an NGS assay are only as good as laboratories’ ability to use their QC data effectively.
With limited time and resources to collect, organize, access, and analyze QC metrics, laboratories may frequently rely on reference materials as binary indicators of Pass/Fail: As long as the expected endpoint results are obtained, an assay is considered to be performing well. The drawback of this approach is that it is reactive, rather than proactive: A sufficient number of failures must occur within a given timeframe before a troubleshooting investigation is performed. By the time a problem is recognized, resources have been wasted and turnaround times (TAT) delayed; in some cases, fidelity of patient results may even have been put at risk. Additional time and costs are then incurred as the investigation proceeds.
Specimen analysis by NGS yields a wealth of information in addition to endpoint variant calls that is indicative of assay performance. Data such as nucleic acid quantity and quality at different steps throughout the workflow (PDF) and sequencing library characteristics are generated every time a reference material is tested. However, these data must be carefully tracked and trended to allow use as highly informative QC parameters. For clinical laboratories whose primary focus is on patient testing and reporting, granular QC metrics may not be captured and reviewed as part of routine test monitoring.
MANY WIDELY-USED QC DATA MANAGEMENT STRATEGIES ARE INSUFFICIENT
In considering NGS laboratories’ many different needs for QC data management, I’m reminded of Steve Jobs’s 2007 introduction of the iPhone. The keynote address he delivered focuses on two primary qualities that are most important for the majority of smartphone users: (1) ease-of-use, and (2) functionality. A very similar approach applies to NGS QC data management; strategies must be intuitive so users have easy access to their data, yet comprehensive to ensure all of the necessary metrics are captured and properly analyzed. Unfortunately, many of the methods currently used are highly manual, prone to error, and result in wasted time and money.
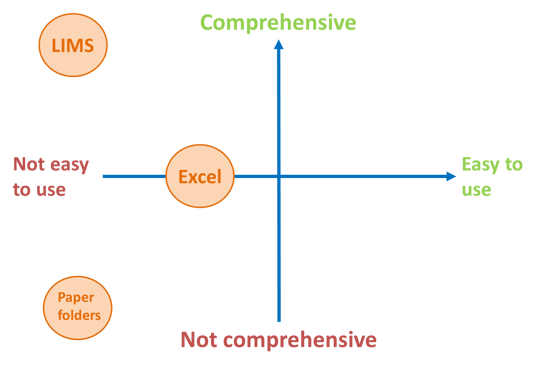
Hardcopy ‘QC folders’ are only suitable for low-throughput production since the document control needs quickly overwhelm as test volume increases. In addition to the obvious physical paperwork burden this method of QC data storage creates, it also discourages effective analysis since viewing results in the proper historic context requires a significant amount of manual effort. It is also highly unlikely to offer thorough traceability since manually recording instruments and reagent lots is time-consuming and distracting for laboratory personnel.
Excel spreadsheets are capable of capturing more information, and allow easier analysis, especially for users who write VBA Macros. This method of data storage is still relatively error-prone, however, and faces significant version control problems when multiple users edit a single file stored on a network. Furthermore, while spreadsheet tools are useful for automated analysis of quantitative data such as variant allele frequencies or sequencing metrics, other types of data such as sample traceability across the NGS workflow must be manually assembled and managed.
Larger clinical laboratories often rely on their Laboratory Information Management System (LIMS) to store QC data. This is a very comprehensive strategy that is capable of capturing data at high resolution, from endpoint calls all the way down to individual laboratory transactions. Unfortunately this vast amount of QC data is stored in a LIMS along with many other types of laboratory and patient data, and must be accessed by personnel who are proficient at writing SQL queries (or a query tool must be developed and validated). After QC metrics have been pulled from the database, they must be formatted and analyzed; for many, Excel is still the best tool available for this downstream work.
Just as Apple ‘reinvented the phone’ to create a device that would sit in quadrant 1 of the ‘ease-of-use versus functionality’ graph, this need also exists in the world of NGS QC data management. Consider the following key features that are essential for any Quality Management System for NGS testing:
QC DATA MANAGEMENT MUST-HAVES
- Metrics must be well organized and easily accessible. Storage in a single location such as a dedicated server guarantees personnel do not have to sort through massive amounts of paper, multiple versions of electronic files, or unrelated data types in order to access their QC metrics. Users should have access to these data via a web browser on any internet-connected computer, without having to install additional software or manage licenses.
- Test performance must be evaluated in real-time using industry-standard methods such as Levey-Jennings plots. This allows trends to be quickly and easily identified, compared to viewing data in tabular format alone. Gradual process drift will be detected earlier thus preventing TAT delays as well as reducing troubleshooting costs.
- Historical context and comparison with a baseline is necessary to discern random noise from significant variation. Because NGS workflows involve a large number of steps that can be affected by variation, it is expected for QC metrics to exhibit a certain amount of random noise, even for the highest quality reference materials. In order to differentiate this normal day-to-day variation from harmful systematic bias, results must be compared with a robust set of historic data. Ideally, statistical analysis should be performed as part of this comparison.
- Traceability is essential for effective troubleshooting. Starting with endpoint results, laboratories must quickly pinpoint instruments and consumables that yield aberrant data. In-process QC metrics such as DNA fragment size or PCR yields must also be quickly and easily accessible.
- QC data management strategies must be adaptable and scalable in order to meet the needs of constantly-evolving clinical tests. As novel targets are added to tests, ideally reference materials should coevolve so they can evaluate performance across the expanded library. Additionally, testing volume increases over time as newly-introduced assays mature, which can mean a greater amount of QC data generated for in-batch controls. Data management methods must be effective throughout all stages of the test lifecycle, not just the initial version that has limited needs.
- Peer review and data sharing are becoming increasingly important as technologies evolve and NGS gains wider adoption in the clinic. Transparency is essential not only for clinical data, but for data derived from widely-adopted standards as well. In fact, testing of the same reference materials is the most effective way to compare assay performance across different sites. QC data that are generated by many different laboratories should be centralized and comprehensively analyzed so each participant gains insights into the performance of their test. Eventually, sufficiently robust sets of community data could enable cloud-based QC services for laboratories who wish to benchmark against metrics that are more comprehensive than their own internal historic data.
- There are unique requirements for managing validation data sets for NGS tests. In addition to typical metrics used for test monitoring, different analytical studies can have unique data management needs. For example, precision studies that evaluate intra-run and inter-run reproducibility need an easy method to check concordance across sample replicates. For studies where certain treatments are applied, a key containing sample ID/ treatment associations must be maintained. Furthermore, different regulatory agencies have different methods of reviewing validation data, as well as different requirements for data retention.
By using these types of QC data management strategies, NGS laboratories can put reference materials to work proactively monitoring their tests. After all, it is not the materials themselves that should grant peace-of-mind, but the effective use of data that they generate.
SeraCare is highly committed to helping clinical laboratories and in vitro diagnostic developers overcome QC challenges they face at both the wet and the dry bench. We understand that the ever-increasing range of NGS applications will always demand ways to ensure test quality. Whether you are developing an assay for somatic mutations, validating a test to predict anti-viral therapy response, or are in production to provide parents with fetal health information, you need a highly effective Quality Management System to ensure patients and doctors receive the best results possible to inform decisions. Pessimists have said, ‘A bad test is as dangerous as a bad drug.’ But why not the glass-half-full perspective? We would prefer to say that a well-characterized, thoroughly-controlled test is every bit as vital to the practice of medicine as a blockbuster therapy.
Miss Clinical Laboratories: You Are Not Alone (Part 1)? Click here to access it now.
Also, to learn more about our iQ NGS Management Software, download our on demand recording orginally presented at the AMP Annual Meeting.
If you would like to learn more about how SeraCare can help fulfill your quality control needs for Precision Medicine, please visit us online, or contact us here or via the comments below.




