Tumor profiling assay workflows start from a tissue biopsy sample that is formalin fixed and paraffin embedded (FFPE), a process that introduces various kinds of damage in the nucleic acids in the tissue specimen. Reference materials that closely mimic the damage profile of patient FFPE samples are lacking. Depurination, depyrimidination, deamination, oxidation, nicks, and double strand breaks may be found in DNA extracted from FFPE tissue, despite the use of extraction kits that attempt to repair some of this damage. We have developed a formalin-damaged, multiplexed biosynthetic reference material, Seraseq® Compromised FFPE Tumor DNA, as well as a companion wild-type (WT) material, Seraseq Compromised FFPE WT, to mimic the damage found in patient samples that can be used to assess the entire tumor profiling workflow. We compared the performance of these reference materials in downstream assays to that of FFPE material with minimal DNA damage such as the Seraseq FFPE WT (DNA/RNA) RM product.
Reference Material Manufacture
Seraseq Compromised FFPE Tumor DNA materials were made by engineering the reference cell line GM24385 with biosynthetic DNA covering 34 variants, incorporating all variant types – SNVs, INDELs, CNVs, and SVs (Table 1). Engineered cells were diluted with WT cells to achieve desired allele frequencies and copy numbers as determined by digital PCR. This mixture of cells, as well as WT cells alone, were either processed with a proprietary FFPE protocol that introduces moderate damage or with a light fixation FFPE protocol.
Table 1. Variants found in Seraseq Compromised FFPE Tumor DNA Reference Material.
Yield and Quality Comparison
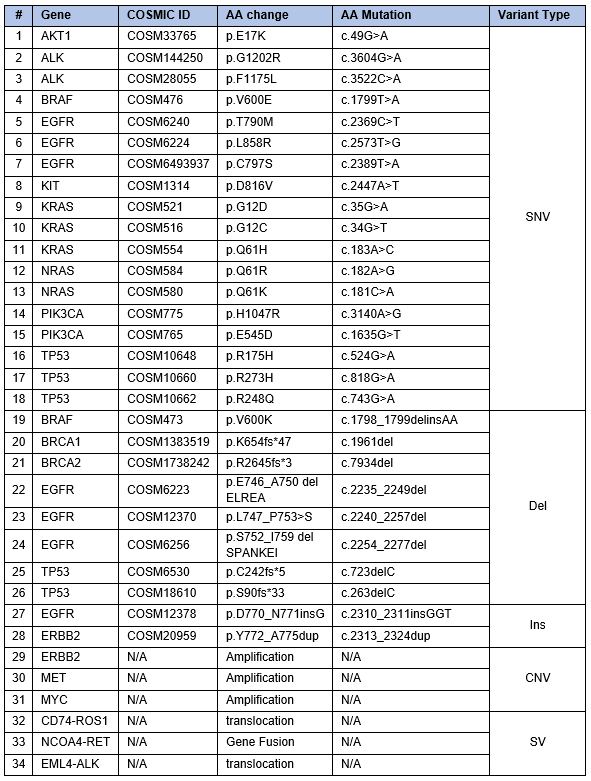
Various FFPE extraction kits were used to isolate DNA and/or RNA from 10 µm sections of each product (Table 2). Yields were similar and sufficient for downstream processes regardless of the method used for fixation.
Table 2. DNA and RNA yields from Seraseq Compromised FFPE reference materials using commercially available extraction kits.
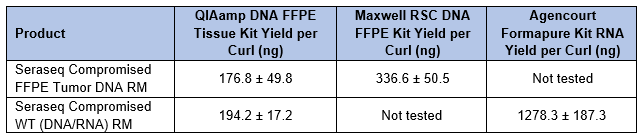
The DNA quality of extracted nucleic acids were assessed using the Agilent TapeStation. The quality of the DNA extracted using the QIAamp DNA FFPE Tissue Kit from each type of block was assessed using the TapeStation Genomic DNA Kit, which calculates the DNA Integrity Number for each sample on a range from 1 to 10, where a score of 10 indicates intact gDNA. The moderate fixation process used for the Compromised FFPE Tumor DNA materials results in DIN values that are 28.9% lower (average DIN value of 3.98 ± 0.27 versus 5.60 ± 0.14) than DIN values of the DNA extracted from FFPE curls made using the light fixation process (Figure 1). Similarly, Compromised FFPE WT materials had lower DIN values than their non-compromised counterparts (average DIN value of 3.95 ± 0.17 versus 5.87 ± 0.17, a 32.7% reduction) as seen in Figure 2.
These data indicate we successfully modified the amount of damage in the material. The average DIN value of our moderate fixation material is close to the average DNA extracted from a patient sample using the same extraction kit results in a DIN value of 3.31.
RNA quality was low for both methods of FFPE manufacture, with measured RIN scores ~1.0 for material extracted from both Seraseq Compromised WT and Seraseq FFPE WT curls with the Agencourt Formapure Kit as measured with the Agilent Bioanalyzer RNA 6000 Nano Kit. These values are similar to RIN values observed for RNA extracted from patient FFPE (average 1.7 ± 0.5) using the same kit2.
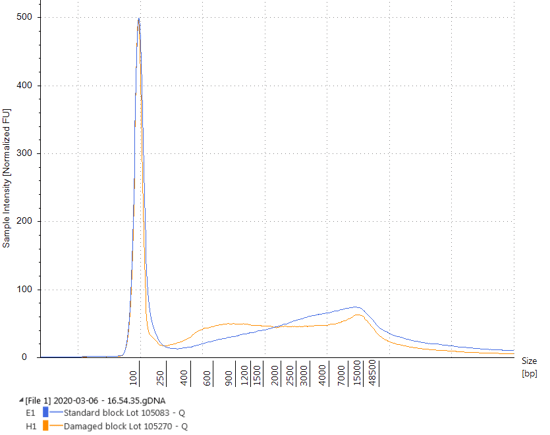
Figure 1. Electropherogram of DNA extracted from Seraseq Compromised FFPE Tumor DNA RM blocks (orange) made with a moderate fixation process and non-compromised FFPE blocks (blue) made with a light fixation process. The moderate fixation process results in less intact genomic DNA (~15 kb) and an accumulate of lower molecular weight DNA.
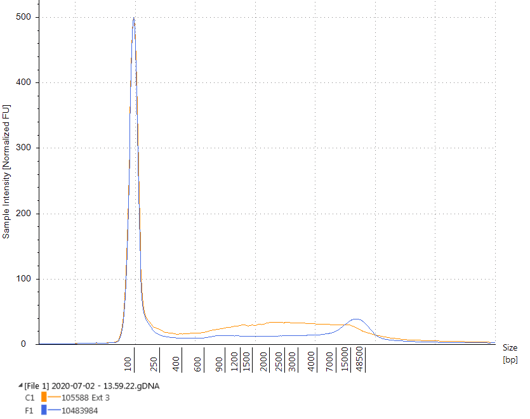
Figure 2. Electropherogram of DNA extracted from Seraseq Compromised FFPE WT RM blocks (orange) made with a moderate fixation process and Seraseq FFPE WT RM blocks (blue) made with a light fixation process. The moderate fixation process results in less intact genomic DNA (~15 kb) and an accumulate of lower molecular weight DNA.
Variant Analysis
When variant analysis was performed with the Illumina TSO500 assay, differential VAFS between DNA extracted from the same Seraseq Compromised FFPE Tumor DNA block with different FFPE extraction kits was observed (Figure 3). This variation is likely due to both the variability in extraction efficiency of the genomic DNA and biosynthetic variants added as well as differences in biochemical activity and bias in the reversal of formaldehyde damage during DNA isolation with the respective kits. Variation in apparent VAF due to extraction method is a known phenomenon1. All variants were detected between 2% and 55% . Similarly, the copy numbers measured for each amplified gene were variable (Figure 4), but well supported in each sample. Similar results were seen when the extracted DNA was analyzed by digital PCR (data not shown).
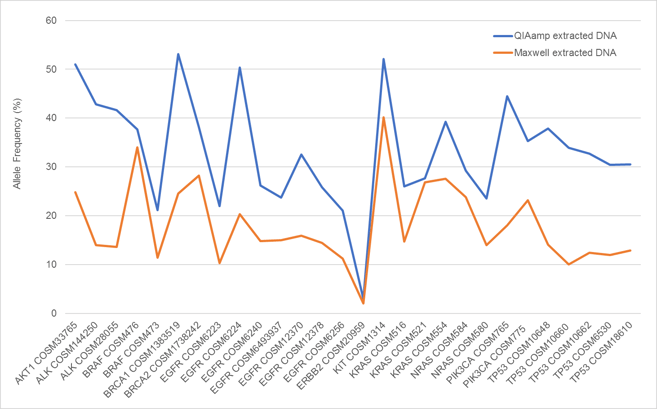
Figure 3. Variant allele frequencies as measured by the TSO500 NGS assay in DNA extracted from Seraseq Compromised FFPE Tumor DNA RM using the QIAamp DNA FFPE Tissue Kit (blue) and the Maxwell RSC DNA FFPE Kit (orange).
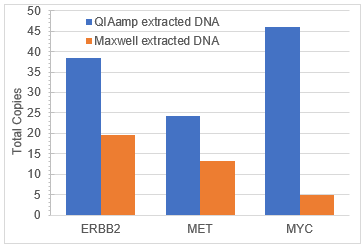
Figure 4. Total copy numbers of amplified genes as measured by the TSO500 NGS assay in DNA extracted from Seraseq Compromised FFPE Tumor DNA RM using the QIAamp DNA FFPE Tissue Kit (blue) and the Maxwell RSC DNA FFPE Kit (orange).
As expected, both DNA and RNA sequencing using the ArcherDx VariantPlex Solid Tumor Assay and the ArcherDx FusionPlex Solid Tumor Assay, respectively, revealed no pathogenic variants in either the Seraseq Compromised FFPE WT Reference Material or the Seraseq FFPE WT Reference Material when samples were sequenced to a depth of ~1 million reads per sample. While average fragment lengths were shorter for the compromised material than the light fixation material (178.0 bp vs 196.8 bp for DNA and 146.9 bp vs 159.5 bp for RNA), both materials performed well in the assays, with sufficient library concentrations (>6 nM for DNA, >26 nM for RNA), high mapped and aligned reads (>99% for both DNA and RNA), and suitable on-target reads (>98% for DNA and >94% for RNA).
Overall, both FFPE methods result in reference materials that are compatible with PCR and NGS (amplicon- and hybrid capture-based) assays. With compromised products, performance can be impacted by the FFPE extraction method deployed to recover amplifiable DNA for downstream analysis. Thus, the Seraseq Compromised Tumor DNA Reference Material will serve well as a qualitative product for assessing the ability of an assay to detect the included variants.
The moderate fixation method more closely mimics archived tissue biopsies of cancer patients while maintaining multiplexing capability and downstream performance making these Seraseq products applicable in NGS assay development, bioinformatics pipeline optimization, and as full process references in clinical NGS assays used for patient testing.
References:
1. Bonnet E, Moutet M-L, Baulard C, et al. Performance comparison of three DNA extraction kits on human whole-exome data from formalin-fixed paraffin-embedded normal and tumor samples. PLoS One. 2018; 13 (4); e0195471.
2. Kresse SH, Namløs HM, Lorenz S, Berner J-M, Myklebost O, Bjerkehagen B, and Meza-Zepeda LA. Evaluation of commercial DNA and RNA extraction methods for high-throughput sequencing FFPE samples. PLoS One. 2018; 13 (5); e0197456.




