This is Part 1 of a 2-part blog reviewing the Genomic Testing webinar and panel discussion featuring Dara Aisner, MD PhD, George Green, PhD and Greg Tsongalis, PhD. With so much rich information, we will be posting two blogs. Part 1 will cover select important themes discussed by each speaker, and Part 2 will review the audience Q&A.
Recently, I had the pleasure of participating in a webinar co-sponsored by LGC SeraCare and GenomeWeb. The topic was “Genomic Testing to Support New Therapies for Advanced Cancer”.
This was an opportunity for all of us to hear from three thought leaders (Dr. Dara Aisner, Dr. George Green and Dr. Greg Tsongalis) who have contributed to the steady adoption of NGS but who also know about the many technical and clinical challenges we still face to ensure all cancer patients have the right diagnosis and best available treatment. The webinar format allowed for presentations by each panelist followed by discussion and audience Q& A.
Dr. Dara Aisner, MD PhD. was the first presenter. Dr. Aisner is a board-certified anatomic and molecular genetic pathologist and clinical informaticist. She is an associate professor of pathology at the University of Colorado School of Medicine in Denver, Colorado and a member of the University of Colorado Comprehensive Cancer Center. She has been a member of numerous oncology pathology and guidelines committees, including the CAP/AMP/IASLC and National Comprehensive Cancer Network NCCN.
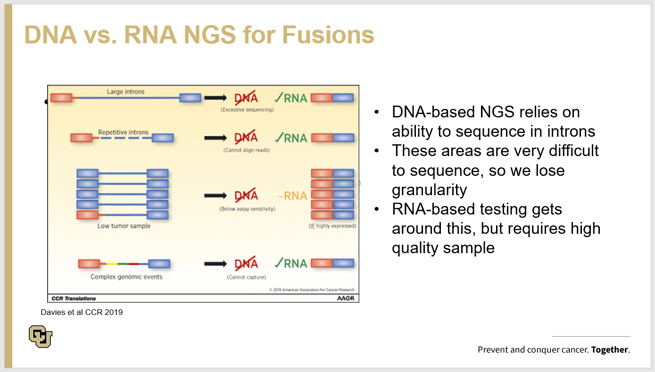
During her presentation, Dr. Aisner used a lung cancer case study to show how important it is to ensure the primary tumor is used to exhaustively test for RNA fusions if the driver mutation is not identified by NGS DNA analysis. The case study showed how a 30 gene NGS panel failed to identify the driver mutation and the patient was selected for immunotherapy based on PD-L1 IHC but unfortunately the patient’s cancer progressed after 6 months. After progression, an NGS RNA workflow did identify a ROS1 fusion and ROS1 inhibitor drug was initiated and the images post anti-ROS1 treatment clearly showed tumor clearance. The chart presented below shows the technical deficiencies detecting pathogenic fusions by DNA alone and why RNA workflows are preferred. Dr. Aisner also pointed out that in both workflows, there is no way around a problem with limited sample or insufficient yield of extracted nucleic acid. For more information on her work, please check out this reference which details advantages detecting RNA fusion with a finite amount of tumor sample. (1)
The next presentation was by Dr. George A. Green. Dr. Green is Executive Director and Head of Pharmacodiagnostic Sciences - Translational Medicine at Bristol Myers Squibb. He has played a key role in establishing pharmacodiagnostics as a core capability within BMS. Dr. Green led development programs in oncology, neuroscience, cardiovascular, and virology, as well as early diagnostic development programs across multiple therapeutic areas and technology platforms.
At the webinar, Dr. Green highlighted the importance of liquid biopsy-based (ctDNA) minimal residual disease (MRD) monitoring to advance patient selection into clinical trials and for early treatment decisions. Dr. Green presented ctDNA tracking data from published clinical studies and it was quite striking to see that if tumor DNA is detectable in blood after initial cancer treatment, it is highly indicative of disease progression.
Dr. Green also listed 12 on-going clinical trials sponsored by multiple organizations where ctDNA MRD assays are in ongoing prospective trials assessing ctDNA Levels with treatment response in early-stage diseases. One CRC phase III trial listed was MGH 18-357 “Early Identification and Treatment of Occult Metastatic Disease in Stage III Colon Cancer”. I was curious to learn more about the trial and the ctDNA indication. Here is the trial description. https://clinicaltrials.gov/ct2/show/NCT03803553 and the following statement is quoted directly from the study.
“After diagnosis and surgical removal of tumors, individuals with metastatic colorectal cancer commonly receive what is called adjuvant chemotherapy treatment, commonly utilizing treatment plans called FOLFOX, CAPOX, or therapy with 5-Fluorouracil.
If all the cancer is not killed, the investigators may be able to detect tumor in the blood called circulating tumor DNA (ctDNA). This is genetic material unique to metastatic colorectal cancer that may be present in the blood stream, and it can be identified through a ctDNA blood test. If ctDNA is present in the bloodstream, it is commonly called micro-metastatic disease (meaning disease that can't be seen detected by CT scans but may be there in the blood). Cancer researchers believe that ctDNA in the blood stream maybe an indicator that cancer is more likely to recur.”
….and from the research component of the trial.
- “the investigators are determining whether there are differences in cancer recurrence in ctDNA positive participants treated with additional therapy versus put on active surveillance.
- determining whether there are differences in health in ctDNA positive participants treated with additional therapy versus put on active surveillance.
- examining whether patients who undergo further therapy experience changes in the ctDNA levels.”
Regarding the genotypic biomarkers, quantitation and limits of detection, the protocol states that all tumors were genotyped, and the primary inclusion genotypic biomarkers are mismatched repair deficiency documented by IHC or protein expression or PCR (MSI-H), blood ctDNA positive and blood ctDNA BRAF V600E mutations are followed. The research aims do include measuring trends to below a detectable threshold value. The investigators selected the LUNAR ctDNA assay from Guardant Health. The analytical sensitivity 95% LoD of this assay has been reported to be 0.3% VAF for 10ng DNA input and < 0.1% VAF for 30ng.(2)
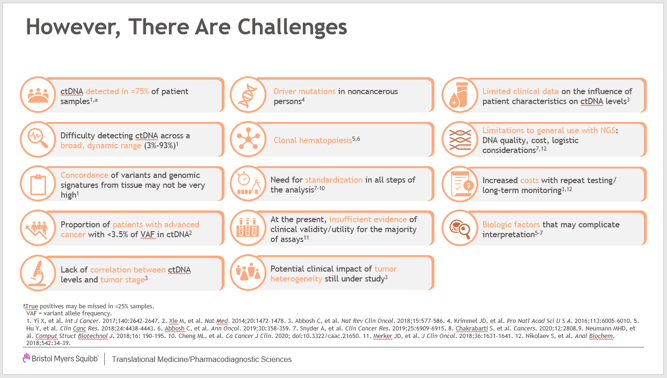
Dr. Green closed out his presentation by providing a chart that showing many challenges we have applying MRD testing. These challenges, as he pointed out, need to be addressed in order to keep advancing MRD for patient care.
The third expert panelist was Dr. Greg Tsongalis. Dr. Tsongalis is the Director of the Laboratory for Clinical Genomics and Advanced Technology (CGAT) at the Dartmouth-Hitchcock Medical Center and Norris Cotton Cancer Center (NCCC). He is also a Professor and Vice-Chair for Research in the Department of Pathology and Laboratory Medicine at the Audrey and Theodor Geisel School of Medicine at Dartmouth. Dr Tsongalis is a past President of the Association for Molecular Pathology (AMP).
Dr. Tsongalis and his team have many years of experience and an outstanding reputation bringing new molecular technologies into the clinical lab through rigorous validation, while never compromising quality. Consequently, I was quite interested in his experience assessing and applying NGS gene panels to patient care. Greg reported that his lab used a 50-gene cancer hotspot panel for 6 years and due to new driver genes being clinically validated and new treatments approved they validated and adopted an ILMN TST170 gene panel in 2020. After just 6 weeks deploying the 170 gene panel to support clinical patient testing, the results were quite impressive. The lab sequenced 25% more tumors than would have been sequenced by the 50 gene panel, detecting an additional 12 fusions, 3 MET ex14 skipping mutations, 1 AR-V7 and 1 EGFR vIII mutations by RNA analysis, and detected >20 variants including 4 CNV amplifications by DNA analysis.
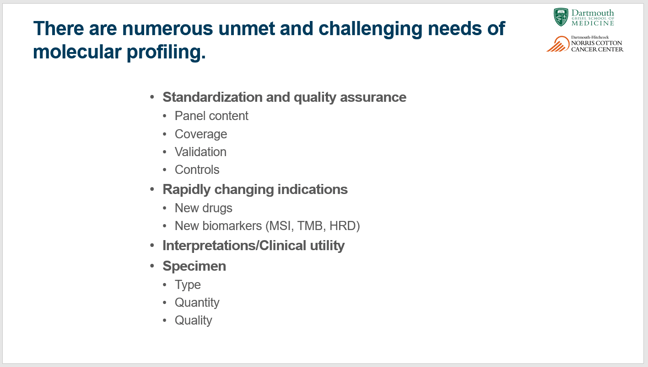
Similar to Dr. Green’s charge to all of us involved in clinical genomics, Dr. Tsongalis listed the many challenges molecular laboratories are faced with today. See the chart below.
In addition to the challenges listed above, Dr. Tsongalis has responded to the consistent request from Tumor boards to help reduce turnaround time (TAT) reporting genotypic results to shorten the period from diagnosis to treatment. For those who are interested to learn more about this topic, please check out Tsongalis et al. (3) Here they assessed CRC-targeting NRAS, KRAS and BRAF mutations. The study compared a laboratory’s standard of care approach (Sanger sequencing or NGS or qPCR) to the fully automated Biocartis Idylla platform for 780 cases to determine test result reporting TAT compared with standard assays. PCR had an average testing TAT of 5.6 days, send-out PCR was 22.5 days, in-house Sanger sequencing was 14.7 days, send-out Sanger was 17.8 days, in-house NGS was 12.5 days, and send-out NGS was 20.0 days. Standard testing had an average turnaround time of 11 days. Idylla averaged TAT of 4.9 days.
I concluded the presentation part of the webinar by reviewing the NGS reference materials and standards we develop and manufacture at LGC Seracare for clinical oncology. These includes tumor profiling, liquid biopsy and immuno-oncology. I also used this opportunity to introduce a number of new Seraseq Oncology products recently released:
Immuno-Oncology:
-
-
- Seraseq Blood TMB, scores 7,13,20,26 at 2%, 0.5% and 0% for assay development, validation and QC
-
MRD:
-
-
- Seraseq ctDNA MRD Panel Mix, consists of tumor-normal matched cell line plus 25 DNA constructs of clinically relevant variants across 15 genes, fragmented, sized and blended at tumor fractions of 0.5%, 0.05%, 0.005% and 0% for ctDNA NGS MRD assay development, validation, QC and PT/EQA
-
Tumor Profiling:
-
-
- Seraseq Lymphoma FFPE and gDNA formats, SNVs and del across BCL2, BRAF, DNMT3A, EZH2, IDH2, MYD88, CXCR4, NOTCH1,2, RHOA, SF3B1, STAT3, STAT5B, TP53, 5-15% VAF
- Seraseq Whole Transcriptome RNA Seq mix, multiplexed to 22 RNA fusion detection by transcriptome RNA Seq NGS assays.
-
For more information about these products please check out www.seracare.com,
You may access the webinar replay here, and stay tuned for part 2 that summarizes the questions and answers.
References:
- Herbst RS, Aisner DL, Sonett JR, Turk AT, Weintraub JL, Lindeman NI. Practical Considerations Relating to Routine Clinical Biomarker Testing for Non-small Cell Lung Cancer: Focus on Testing for RET Fusions. Frontiers in medicine. 2020;7:562480.
- Artieri C, et al Analytical validation of a tissue agnostic ctDNA MRD assay using tumor specific methylation and somatic variant profiles in early-stage CRC. ASCO Annual Meeting. 2020.
- Tsongalis GJ, Al Turkmani MR, Suriawinata M, Babcock MJ, Mitchell K, Ding Y, et al. Comparison of Tissue Molecular Biomarker Testing Turnaround Times and Concordance Between Standard of Care and the Biocartis Idylla Platform in Patients With Colorectal Cancer. American journal of clinical pathology. 2020;154(2):266-76.




