 Labs that demonstrate best practice clinical next-generation sequencing (NGS) quality management programs utilize positive run controls, designed for this purpose, to monitor assay analytical performance across each step of the clinical NGS workflow. Common parameters for assessing the analytical performance of a clinical NGS run include sensitivity (true positives), specificity (true negatives), and limit of detection (LOD) for biomarkers of interest. If the quality control material does not perform as expected the run is failed, troubleshooting begins until the problem is solved, and the sample batch is retested.
Labs that demonstrate best practice clinical next-generation sequencing (NGS) quality management programs utilize positive run controls, designed for this purpose, to monitor assay analytical performance across each step of the clinical NGS workflow. Common parameters for assessing the analytical performance of a clinical NGS run include sensitivity (true positives), specificity (true negatives), and limit of detection (LOD) for biomarkers of interest. If the quality control material does not perform as expected the run is failed, troubleshooting begins until the problem is solved, and the sample batch is retested.
As part of their QC program, labs also look at run statistics coming from their sequencer and bioinformatics pipeline to ensure the data fall within expected ranges and distributions. This combination of monitoring analytical performance from positive run controls as well as sequencing run statistics for all samples provides increased confidence in results.
However, some laboratories use sequencing run data as a substitute for positive run controls. Our analysis of data from a recent industry-wide survey of clinical NGS QC practices and a study of over 150 NGS runs reveals some concerning issues that could occur when this takes place. Additionally, our analysis uncovers how labs may save money by leveraging positive run controls to reduce costs associated with excessive downtime related to troubleshooting sources of assay failure.
False Positives and Negatives May Occur Even When NGS Run Metrics Pass
In a dataset of over 150 clinical NGS runs covering 1.5 years that included a multiplexed positive control in each run, 24 metrics were tracked including statistical measures from the sequencing runs and the bioinformatics pipeline. Many of these metrics appear in the recommendations from CAP/AMP. Each metric had clearly established acceptable and warning ranges that were derived from the validation study performed.
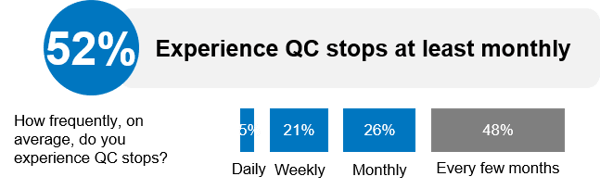
11% of these runs were deemed failed runs as a result of either false positives or false negatives being present in the control material and occurred on average once per month. This tracks in line with the majority of respondents who reported experiencing a QC stop at least monthly in the clinical NGS QC practices survey we performed in partnership with GenomeWeb (Figure 1).
Figure 1. Data from Clinical NGS QC Practices Survey Shared During “Trends in Clinical NGS QC Management” Webinar. Source: Trends in Clinical NGS QC Management: Results from a Practitioner Survey. September, 2018.
As we dig deeper into the data we find that 30% of those failed runs had no warning or failures in the 24 sequencing metrics, meaning from a practical point of view these failures were “silent” and the only safeguard was the use of positive run controls.
Visualizing the data with a Levey-Jennings plot shows how run failures can occur well within the two standard deviations that labs typically use to flag results that require further investigation. A representative example of this is seen using the mean read depth (Figure 2).
Figure 2. A study comprising of >150 NGS runs resulted in 11% false positives or false negatives (red circles). 30% of those runs with false positives or false negatives did not have a failed mean read depth (defined as greater than 2 SD) and were only detected using a positive control (red circles within 2 SD)
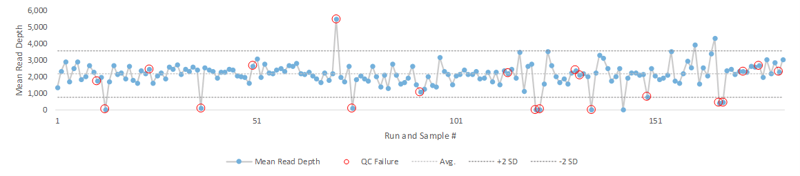
These silent failures would be difficult to detect without a positive run control and the ground truth dataset it represents. This value is particularly pronounced in some applications of oncology testing where low-level allele frequencies are critical such as liquid biopsy and offer additional assurance when reporting out clinical results.
Positive Run Controls May Reduce Downtime
Run failures often trigger troubleshooting efforts and the ability to get the assay back online quickly makes a material difference on the lab’s ability to maintain their targeted turnaround time and minimize any financial losses related to potential test send outs or spent consumables. As we discussed above, the majority of labs running clinical NGS assays are faced with a QC stop on a monthly basis and therefore must make the most use of the tools available to them to get back up and running as quickly as possible.
One such tool is the consistent longitudinal data generated by a positive run. In the survey data we see a distinct difference between a labs’ ability to get back up and running quickly depending on their positive run control usage. Out of the respondents in the survey mentioned earlier, 63 oncology testing labs who routinely used positive run controls became operational within three days after a QC stop had occurred compared to 18 who used positive run controls more infrequently. This strongly suggests that the use of positive run controls and the data generated by that use can be leveraged to more quickly resolve sources of run failure and minimize downtime.
It should also be noted that not all positive controls are created alike. A commercially available material that is demonstrated to be repeatably manufactured offers considerably more utility and confidence to a lab as they run assays and longitudinally track and trend data over time in comparison to remnant samples that once depleted are replaced as well as are subject to degradation as they go through multiple freeze/thaw cycles.
With this analysis of both a comprehensive set of clinical NGS run data as well as a representative survey of clinical NGS QC practices, it is clear that there are risks inherent to quality management programs that rely heavily on the statistical measures offered by sequencing instrumentation and bioinformatics pipelines without ground truth datasets represented by positive controls. Additionally, routine use of positive controls may reduce downtime which in turn reduces costs of sequencing reagents spent troubleshooting run failures and test send outs.
Another often overlooked fact is that LDTs use research use only (RUO) labeled reagents, solutions, instruments, and software thus the testing lab, not the manufacturer of these RUO materials, is responsible to identify assay performance changes and historical experience says that control data is the most effective way to monitor changes.
Managing and analyzing the data that is tracked and trended for controls is a key aspect to implementing this robust approach to quality management, and you can click here to learn more about how SeraCare’s iQ NGS QC Management solution helps labs jump start their QC management programs by offering CAP/AMP metric templates and how the solution tightly integrates with the Seraseq line of reference standards designed specifically for NGS.
Purpose-designed NGS quality control materials and QC software are tools Major Tom can rely on to keep the spaceship flying.
 How Can Labs Implement a Best-In-Class, Robust, and Compliant Next-Generation Sequencing QC Program?
How Can Labs Implement a Best-In-Class, Robust, and Compliant Next-Generation Sequencing QC Program?
Two-Part Workshop Video: Two scientific thought leaders discuss applying up-to-date quality control systems and standardization to NGS oncology assays to meet current guidelines and regulatory compliances.




