Microsatellites are simple tandem repeats that are present at millions of sites in the human genome. Microsatellite Instability (MSI) is defined as a change of any length due to either insertion or deletion of repeating units in a microsatellite within a tumor compared with normal tissue.1 The molecular mechanism for the change in repeat length is slippage of nascent DNA strand with respect to the template strand during replication followed by failure to recognize the mismatch due to deficiency in mismatch repair genes.
Mismatch repair (MMR) proteins include MLH1, MSH2, MSH6, and PMS2. These proteins are stable in cells only as heterodimers. Immediately after replication, the MutS complex (comprised of MSH2 and MSH6) recognizes the small loops indicative of mismatches. MutS recruits the MutL complex (comprised of MLH1 and PMS2) to assemble close to the mismatch, nick the DNA, and initiate repair of the mistake.2 Deficiencies in mismatch repair can be a result of inherited changes in the MMR proteins (Lynch syndrome) or due to acquired mutations or silencing of these proteins within a tumor (somatic changes).
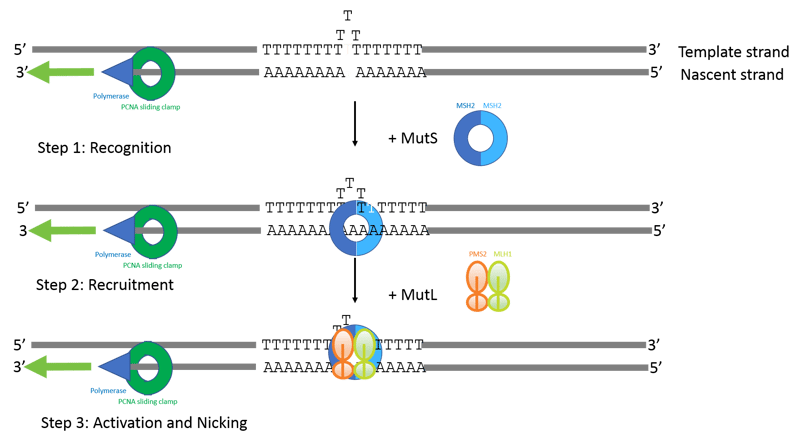
Figure 1: Model of Mismatch repair adapted from references 2 and 3. The example shows a homopolymer of 18 Ts on the template strand that have been incorrectly replicated to generate only 15 as on the newly replicated strand. The presence of the sliding clamp PCNA, along with the nicks used to load the sliding clamp help to identify the nascent strand. MutS complex recognizes the small loop indicative of a mismatch and then undergoes an ATP-dependent conformational change which allows the MutL complex to bind. The MutS/MutL complex is thought to be able to translocate along the DNA, interact with PCNA, and induce nicking near the mismatch site. The nicks then recruit exonucleases and other proteins to remove and repair the mismatch.
The first NCI-sponsored consensus conference on MSI was held in Bethesda, MD in 1997. From this meeting, a reference panel was proposed which came to be known as the Bethesda panel and included 2 mononucleotide markers and 3 dinucleotide markers. The markers serve as functional indicators of deficiencies in mismatch repair (dMMR) and categorization of tumors as MSI-high, MSI-Low and MSI-stable based on these markers was recommended1. Later, it became clear that dinucleotide repeats are less sensitive than mononucleotide repeats for detecting MSI-high4, and the Bethesda panel was updated accordingly to include 5 mononucleotide repeats.
There has been increased focus on identifying MSI-high tumors because of the correlation of MSI status and response to immunotherapy. Looking across twelve different cancer types, Dung Le and colleagues showed that cancers with MMR deficiency were sensitive to immune checkpoint blockade with anti-PD-1 antibodies5. Eighty-six patients with advanced, MSI deficient cancers were enrolled in the KEYNOTE-016 study and treated with the anti-PD-1 antibody pembrolizumab. Objective radiographic responses were seen in 53% of cases and 21% of cases achieved complete radiographic response. The authors performed deep sequencing of the CDR3 regions of the T-Cell Receptor gene (TCRseq) from three of the responding patients. They identified clones that were present at very low frequency (often undetectable) in the peripheral blood before pembrolizumab treatment, but had rapidly increased after initiating treatment. For one of these patients, they went on to functionally characterize the T-cell clone’s ability to bind mutant peptides. They tested the patient’s post-treatment peripheral blood for reactivity against the 15 top candidate mutation-associated neoantigens (MANAs) and found that some of the T-cell clones that expanded in response to therapy were specific for these mutation associated neoantigens. The study is significant because it provided supporting data for the hypothesis that the large number of mutant neoantigens in MMR-deficient cancers make them sensitive to immune checkpoint blockade. This trial and others eventually led to the FDA approval of pembrolizumab (Keytruda®) in May 2017: the first FDA approval of a drug based on the presence of a particular biomarker (MSI/dMMR) irrespective of tissue origin.
There are many ways that deficiencies in mismatch repair and the resulting microsatellite instability can be detected in the lab. IHC, targeting MLH1, MSH2, MSH6 and PMS2 is performed in many labs because of the familiar workflow. However, a proportion of mutations in MMR genes (1-5%) have a missense mutation that results in functional deficiency, but the protein retains immunogenicity and thus leads to false negatives by IHC6. PCR and capillary electrophoresis fragment analysis-based testing using Promega’s MSI Analysis System v1.2 has 5 mononucleotide targets plus 2 controls has been the gold standard and was used in the KEYNOTE-016 trial. The advantage of the PCR based MSI analysis system over IHC is that they are functional-based tests. As reported in the AMP 2017 meeting6, new PCR-based testing systems are in development (Promega MSI Analysis System v2.0) that use a panel of long mononucleotide repeats (52 to 60 base pairs long) as well as 4 of the 5 original markers from the original test kit (BAT-25, BAT26, NR-21 and Mono-27). Longer repeats may be more sensitive to deficiencies in mismatch repair7.
Increasingly, NGS-based MSI testing is being offered at a number of laboratories, including the OmniSeq MSI NGS test, FoundationOne CDx test and MSK-IMPACT test. The primary advantage of these NGS-based MSI detection assays is that they evaluate hundreds or even thousands of MSI loci, rather than being limited to the 5 loci assessed in traditional PCR-based tests. Additionally, assessing MSI using a “stand-alone” assay would be inefficient to perform on every cancer patient since only about 5% of cancers are MSI-high; however, NGS allows MSI detection as part of a more comprehensive analysis that includes assessment of driver mutations for targeted therapy and total tumor mutation burden. Another advantage of NGS-based MSI testing is that it does not require tumor-normal comparison, which is a major advantage for clinical labs8.
No matter the testing method, clinical testing for MSI has challenges. PCR based tests screen a limited number of functional microsatellite markers (>10 sites in the genome). Changes can be subtle, and not present at the majority of loci tested6. For example, Yang Wang and colleagues showed that the mean shift in the five mononucleotide repeat markers: BAT25, BAT26, NR21, NR24, and MONO27 was only 3 nucleotides in endometrial cancer and that 76% of the time, multiple markers had shifts of only 1 nucleotide9. Additionally, changes can be tumor type specific10; for example, the genes frequently targeted by MSI in CRC genomes rarely harbor DNA slippage events in Endometrial Cancer genomes11. Therefore, testing only a very limited number of markers increases the risk of false negatives.
NGS may be preferable because it examines more microsatellite markers than PCR and may increase the sensitivity of the test for non-CRC samples12, 13. However, sensitivity at low allele frequencies may be limiting. Sequencing the MMR genes themselves is also challenging. Pseudogenes of PMS2 can present challenges. As shown by Talseth-Palmer and colleagues, obtaining desired effective read depth across the MMR genes also can be difficult. Large deletions may not be effectively called by some bioinformatics pipelines.
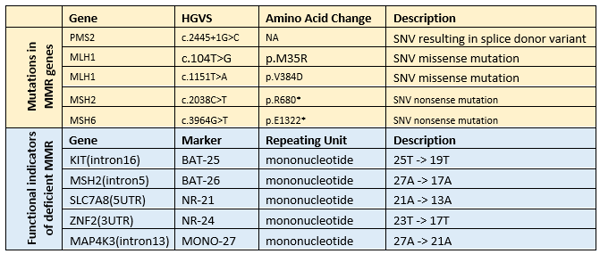
Well characterized reference materials can ease the testing challenges and give greater confidence in the results from both PCR-based and NGS-based tests. For PCR-based tests, they can help standardize the interpretation of fragment analysis electropherograms by different reviewers. For NGS-based tests they can help tune the bioinformatic pipeline and ensure run-to-run consistency. SeraCare’s VariantFlex library now contains both mutations in the key MMR genes as well as altered lengths of the most commonly assessed microsatellite loci (Table 1). Reference materials are offered in a customizable format that allows development, optimization, and bioinformatics tuning for either PCR or NGS-based test systems.
For more information on custom materials including key MMR genes and other key altered lengths for development, optimization, and tuning visit our VariantFlex NGS library page.
Download this poster to see how the VariantFlex technology allows rapid configuration of reference samples to meet clinical laboratory needs for cancer patient analysis of somatic cancer mutations, copy number amplifications, DNA-based structural variants, RNA-based gene fusions, and germline mutations.
References:
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998 Nov 15;58(22):5248-57.
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology.Annu Rev Biochem. 1996;65:101-33.
- Kunkel TA, Erie DA. Eukaryotic Mismatch Repair in Relation to DNA Replication. Annu Rev Genet. 2015;49:291-313. doi: 10.1146/annurev-genet-112414-054722.
- Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004 Feb 18;96(4):261-8.
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, Wong F, Azad NS, Rucki AA, Laheru D, Donehower R, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Greten TF, Duffy AG, Ciombor KK, Eyring AD, Lam BH, Joe A, Kang SP, Holdhoff M, Danilova L, Cope L, Meyer C, Zhou S, Goldberg RM, Armstrong DK, Bever KM, Fader AN, Taube J, Housseau F, Spetzler D, Xiao N, Pardoll DM, Papadopoulos N, Kinzler KW, Eshleman JR, Vogelstein B, Anders RA, Diaz LA Jr. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. 2017 Jul 28;357(6349):409-413.
- Eshleman JR. “MSI as a Research Marker for Immunotherapeutic Response”. Association for Molecular Pathology Annual Meeting, Nov. 15, 2017, Salt Palace Convention Center, Salt Lake City, UT.
- Jeffery W. Bacher, Chelsie K. Sievers, Dawn M. Albrecht, Ian C. Grimes, Jennifer M. Weiss, Kristina A. Matkowskyj, Rashmi M. Agni, Irina Vyazunova, Linda Clipson, Douglas R. Storts, Andrew T. Thliveris, Richard B. Halberg. Improved Detection of Microsatellite Instability in Early Colorectal Lesions. PLoS One. 2015; 10(8): e0132727.
- Vanderwalde A, Spetzler D, Xiao N, Gatalica Z, Marshall J. Microsatellite instability status determined by next-generation sequencing and compared with PD-L1 and tumor mutational burden in 11,348 patients. Cancer Med. 2018 Mar;7(3):746-756.
- Wang Y, Shi C, Eisenberg R, Vnencak-Jones CL. Differences in Microsatellite Instability Profiles between Endometrioid and Colorectal Cancers: A Potential Cause for False-Negative Results? J Mol Diagn. 2017 Jan;19(1):57-64.
- Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. 2013 Nov 7;155(4):858-68.
- Gurin CC, Federici MG, Kang L, Boyd J. Causes and consequences of microsatellite instability in endometrial carcinoma. Cancer Res. 1999 Jan 15;59(2):462-6.
- Salipante SJ, Scroggins SM, Hampel HL, Turner EH, Pritchard CC. Microsatellite instability detection by next generation sequencing. Clin Chem. 2014 Sep;60(9):1192-9.
- Hempelmann JA, Lockwood CM, Konnick EQ, Schweizer MT, Antonarakis ES, Lotan TL, Montgomery B, Nelson PS, Klemfuss N, Salipante SJ, Pritchard CC. Microsatellite instability in prostate cancer by PCR or next-generation sequencing. J Immunother Cancer. 2018 Apr 17;6(1):29.
- Talseth-Palmer BA, Bauer DC, Sjursen W, Evans TJ, McPhillips M, Proietto A, Otton G, Spigelman AD, Scott RJ. Targeted next-generation sequencing of 22 mismatch repair genes identifies Lynch syndrome families. Cancer Med. 2016 May;5(5):929-41.





