“Happy families are all alike; every unhappy family is unhappy in its own way.”

I have always thought the opening of Anna Karenina applies for many things beyond familial harmony (or lack thereof). Certainly, in the world of molecular genetic diagnostics, conclusive results are usually obtained for most patients; however, there are times when a final result is more elusive than conclusive. When this occurs, it may seem as though no two challenges are ever the same.
The following are real examples – presented in general terms for patient and institutional confidentiality – of difficult, unanticipated, and even bizarre cases I encountered during my time in clinical testing for predisposition to hereditary disease. Each of these situations required extraordinary effort, dedicated time, and additional resources for resolution. At the end of each day, satisfaction came from knowing that another problem solved was another patient helped in making life-altering medical management decisions.
The Right Frame of Mind
Mutations that appear in homopolymers are notoriously difficult to detect across different technologies. Enzyme slippage during amplification and alignment issues for Next-Generation Sequencing (NGS) may lead to generation of false results for this type of variant. The importance of thoroughly evaluating performance for mutations in homopolymers was underscored for me by a case involving 12 identical bases that were split up by the presence of a single unique nucleotide in the middle: XXXXXXXYXXXXX, where X represents repeated bases and Y represents the unique base.
For the wildtype allele, this sequence did not present problems. However, a recurrent variant allele was detected which, based on Sanger sequencing data, could have multiple interpretations. Examination of the Y base position alone seemed to indicate the presence of a Y>X substitution; however, there was also secondary sequence present downstream which seemed to suggest a one-basepair frameshift caused by deletion of the Y nucleotide. Was the true nature of this variant a single-base substitution which resulted in a continuous 13-base homopolymer, thus causing enzyme slippage and the presence of an artifactual “stutter” sequence downstream, or was it really a single-base deletion leading to a true “n-1” frameshift? As these two variants may have very different clinical interpretations (and treatment outcomes), accurately differentiating between the two possibilities was vitally important.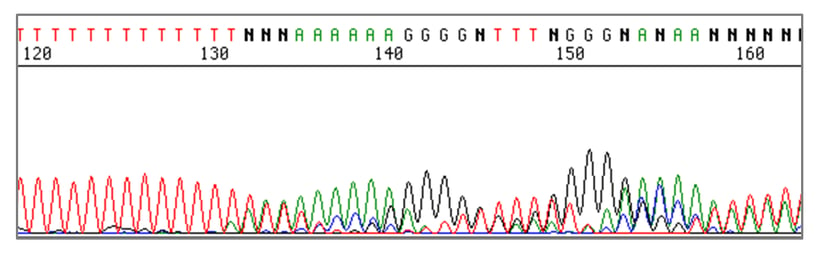
“Stutter” sequence after a homopolymer.
Image courtesy of University of Michigan Medical School.
After much trial and error due to the lack of a well characterized reference capable of establishing assay performance for this difficult region, the variant was eventually confirmed as a single-base substitution by using a high-fidelity enzyme in the genomic DNA amplification step. Clinical interpretation was assigned after sufficient data were collected, and patients were able to receive appropriate treatment.
Equality Is Important
When testing for germline mutations, one of three outcomes is generally expected:
- Variant is not present (wildtype, 0% variant allele frequency)
- Variant is present on one allele (heterozygous, 50% variant allele frequency)
- Variant affects both the maternal and paternal alleles (homozygous, 100% variant allele frequency)
Unfortunately, this is not always the case and detection of variants at less than (or in certain cases, greater than) 50% allele frequency can lead to many questions.
In some instances, the appearance of variants at unequal allele frequency relative to the wildtype can be readily explained by medical history – patients who are suffering from a hematologic malignancy or who have recently received a bone marrow transplant may exhibit this “phenotype” due to a somatic mutation or foreign alleles present in the specimen. Alternatively, the explanation may simply be a technical artifact caused by a variant located at a primer annealing site that disrupts binding, resulting in allelic bias and an apparent misbalance in allele fraction. Unequal germline variants may even indicate the presence of a large rearrangement such as duplication of one or more exons, which could result in a monoallelic variant frequency of 33% (1 variant allele, 2 wildtype alleles) rather than 50%. Finally, true mosaicism is also an underlying cause of unequal variants.
Limit of detection (LOD) is not typically something that is evaluated for germline assays as it is for somatic mutation tests. However, given that Sanger sequencing should in most cases have sufficient sensitivity to detect variants robustly down to approximately 15-20% allele frequency, and NGS-based germline tests should detect variants at even lower frequency, it is important to thoroughly understand the LOD of tests used for inherited disease screening.
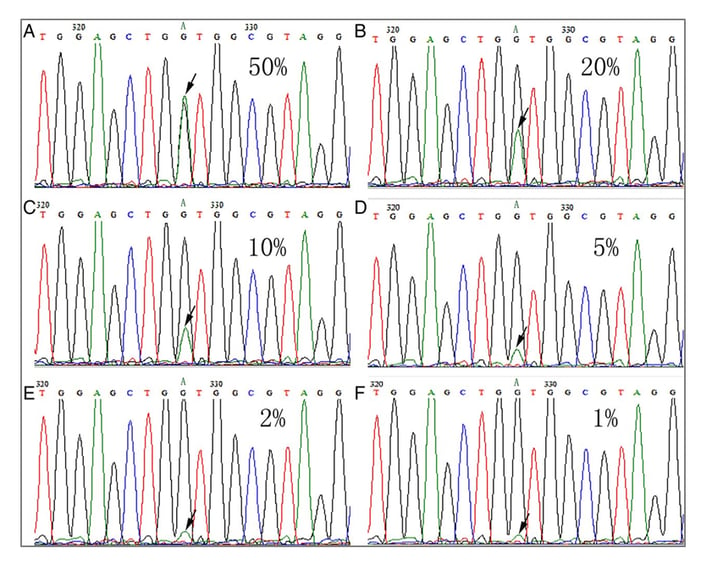
Variants present at 50% allele frequency down to 1% allele frequency on Sanger sequencing.
Gao J, Wu H, Wang L, et al. Validation of targeted next-generation sequencing for RAS mutation detection in FFPE colorectal cancer tissues: comparison with Sanger sequencing and ARMS-Scorpion real-time PCR. BMJ Open 2016;6:e009532. doi: 10.1136/bmjopen-2015-009532.
In fact, it was reasonable to suspect in development that an NGS-based germline panel I helped design would be capable of detecting very low frequency mutations. Indeed, after the test was running in production, such variants were observed; we eventually hypothesized that circulating tumor DNA was the probable source.
Mutations that do not neatly fall into the heterozygous or homozygous bins may or may not be actionable. In some cases, though, the ability to detect variants at unexpected allele frequencies can be critical for downstream decision making.
It’s Wildtype a Deletion an Insertion
Alu elements are common repetitive sequences found throughout the genome which play an important role in disease risk and formation. These transposable elements may insert at functional loci, thereby disrupting native gene function. Because Alu insertions are typically around 300 basepairs in length, they are above the upper size limit for detection by most sequencing assays. These mutations consist of foreign sequence (as opposed to insertion of duplicated sequence at the native locus), meaning they are generally not detected by methods such as array comparative genomic hybridization (aCGH) or multiplex ligation-dependent probe amplification (MLPA®). The result is difficult-to-detect variants that have high clinical severity.
Results across different methodologies for one particular Alu insertion I have observed were as follows:
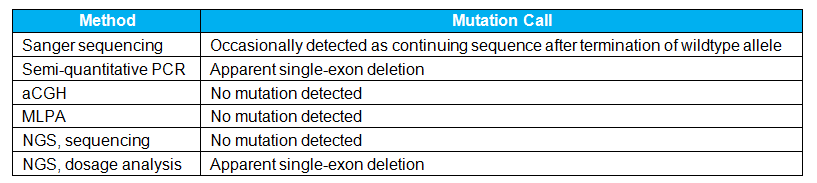
Due to inconsistent and discrepant results across different assays, it was necessary to develop a highly manual test capable of reliably and conclusively confirming the identity of this mutation. This was not easy since the mutation is relatively rare, which made it difficult to obtain a positive control for development, validation, and batch control for the custom assay. However, this test has demonstrated great value since entering production as it has been used to confirm or refute the presence of this deleterious Alu insertion for many patients.
Hindsight is 20/20, but it is safe to say that in each of these unusual cases (and countless others) availability of well characterized reference materials would have offered tremendous scientific and clinical value. Time and resources for resolution would have been reduced, and confidence in patient results would have been bolstered. Unfortunately, it is not always easy to predict the types of issues that will be encountered in high-throughput production testing, which can make proactivity difficult. However, even if we are to accept the argument that inconclusive results are fringe cases which happen so infrequently that it is not worth the time and expense to include a reference material in a process that works for most samples, there are many important efforts for which robust QC tools are essential.
FDA Approval of a Laboratory Developed Test as a Companion Diagnostic
For those familiar with FDA-approved companion diagnostic (CDx) tests such as Roche’s EGFR cobas® test for cfDNA or Foundation Medicine’s FoundationFocus™ CDxBRCA assay, you are most likely aware of the large number of analytical validation studies required. My own experiences in obtaining FDA approval for a Laboratory Developed Test (LDT) as a CDx are no different. In addition to designing validation studies required for a Class III device, writing protocols and reports, coordinating with the lab, and troubleshooting, I also ended up leading a sample procurement effort to try and ensure sufficient access to the astonishing amount of mutation-positive material we needed for these studies. This painstaking effort involved high-touch management of residual patient specimens, reaching out to previously tested patients who were positive for a valuable mutation and were also willing to send in a sample for compensation, and even an internal blood drive. Certainly, not all validations match the scope of a Pre-Market Approval (PMA) submission, but many of the pains associated with obtaining enough material to test are still there.
SeraCare is helping molecular diagnostic scientists and clinicians conserve time and resources, and ensure that patients receive the most accurate and precise results irrespective of test methodology used, specimen type submitted, or where the test is performed. Effective QC tools are not abstract concepts simply used to grant peace of mind; they are real, highly informative materials and practices that are essential for all tests which provide vital information to genetic counselors, physicians, and patients. Testing difficult to detect variants during assay validation studies is one way a lab can effectively challenge their assay to ensure performance. At SeraCare we have great pride in developing and manufacturing reference materials that are rich in difficult to detect variants that prove to the clinical lab directors and to Laboratory Regulators that assay performance meets patient needs.
Please get in touch with us if you are in need of reference materials for your inherited cancer test, cardiomyopathy diagnostic assay, oncology testing, or non-invasive prenatal testing applications.





