The 11th International Symposium on Minimal Residual Cancer was held this month and much of the conference was devoted to new minimally invasive methods for circulating tumor cell enrichment and or the analysis of circulating tumor DNA. Today’s clinical needs are to measure disease burden, track mutations over time, or to detect early resistance and all of these applications require extremely sensitive, robust assays.
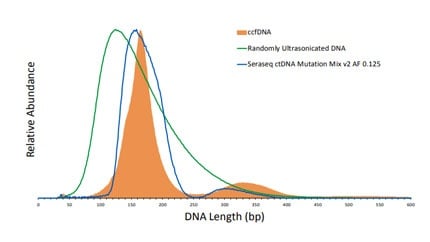 To achieve ctDNA assay requirements that measure at or below 0.1% variant allelic fractions (VAF), matched reference materials that mimic patient samples are required during assay design and testing. SeraCare is collaborating with Drs. Tony Godfrey, Boston University School of Medicine and Anders Ståhlberg, Sahlgrenska Cancer Center University of Gothenburg on ctDNA assays and materials. Drs. Godfrey and Ståhlberg have developed a method called Simple Multiplexed PCR-based barcoding of DNA for Sensitive mutation detection using Sequencing (SiMSen-Seq)[i]. Their method bridges digital PCR for sensitivity and NGS for multiplexing by development of a proprietary library construction that uses reduced primer concentrations, elongated PCR extension times, and hairpin-protected barcode primers. SiMSen-Seq can achieve sensitivities <0.125% (VAF). Seraseq™ technology, from SeraCare, is a biosynthetic technology with wide dynamic range capabilities (0.1-5% VAF) that produces patient-like reference materials containing SNVs, INDELs and CNVs. These quality control reference materials can be used to help test and validate assay sensitivity. At the 11th International Symposium on Minimal Residual Cancer poster session, we reported detection and quantification of four mutations in a Seraseq reference material by SiMSen-Seq as a new approach to achieve ctDNA sensitivity requirements for determining and monitoring MRD.
To achieve ctDNA assay requirements that measure at or below 0.1% variant allelic fractions (VAF), matched reference materials that mimic patient samples are required during assay design and testing. SeraCare is collaborating with Drs. Tony Godfrey, Boston University School of Medicine and Anders Ståhlberg, Sahlgrenska Cancer Center University of Gothenburg on ctDNA assays and materials. Drs. Godfrey and Ståhlberg have developed a method called Simple Multiplexed PCR-based barcoding of DNA for Sensitive mutation detection using Sequencing (SiMSen-Seq)[i]. Their method bridges digital PCR for sensitivity and NGS for multiplexing by development of a proprietary library construction that uses reduced primer concentrations, elongated PCR extension times, and hairpin-protected barcode primers. SiMSen-Seq can achieve sensitivities <0.125% (VAF). Seraseq™ technology, from SeraCare, is a biosynthetic technology with wide dynamic range capabilities (0.1-5% VAF) that produces patient-like reference materials containing SNVs, INDELs and CNVs. These quality control reference materials can be used to help test and validate assay sensitivity. At the 11th International Symposium on Minimal Residual Cancer poster session, we reported detection and quantification of four mutations in a Seraseq reference material by SiMSen-Seq as a new approach to achieve ctDNA sensitivity requirements for determining and monitoring MRD.
The average DNA yield from a 5 ml (125 ng) DNA aliquot of the Seraseq reference material diluted in an artificial human plasma was 110 ng (range 98.4-124.8). 50 ng of DNA which is ~15,000 genome equivalents was used to generate 4-plex libraries KRAS c.35G>A G12D, BRAF c.1799T>A V600E, PIK3CA c.3140A>G H1047, and NRAS c.182A>G Q61R with SiMSen-Seq. The allele detection by SiMSen-Seq is shown in the table below.
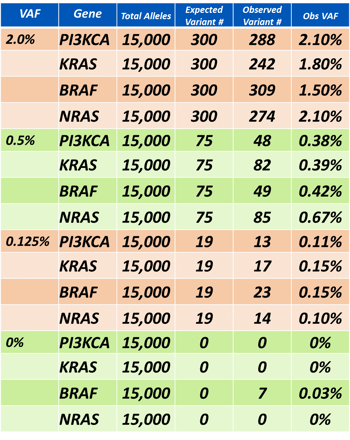
Of special note is that the lowest targeted %VAF (0.125%) represents ~20 variants in 50 ng material and the average of the four variants detected was 16 with a range of 13-23. We concluded from this data set that Seraseq ctDNA technology does provide a well-defined, contrived reference material to evaluate and optimize the liquid biopsy assay process from DNA isolation to absolute sensitivity and SiMSen-Seq appears to be a very accurate and sensitive assay for the detection of known variants down to at least 0.125% VAF and ~ 20 absolute copies. The next steps in our collaborative work with SiMSen-Seq and Seraseq technologies is further optimization and analytical validations to levels less than 0.125% VAF and then applications to minimal disease detection.
[i] Stahlberg A. et al 10.1093/nar/gkw224
To download our poster Application of the Highly Sensitive SiMSen-Seq Assay and Seraseq®-Designed Reference Materials to Minimal Residual Disease Detection, click below.





