Non-invasive prenatal screening (NIPS) is currently offered in over 80 countries, covering over 80 million annual births, with an estimated volume of over one million screening tests performed annually. First offered in 2011, there has been rapid adoption of these genomic tests in the marketplace.
There are many parallels between non-invasive prenatal screening and circulating tumor DNA mutation testing for oncology:
- The analyte, circulating cell free DNA, is shed from a growing organ mass. The placenta or the tumor sheds fragmented DNA within an encapsulated microvesicle into the circulation.
- The fraction of DNA being measured is small thus requiring low limits of detection (LOD). Assay performance needs to be acceptable at 4% fetal fraction in the case of NIPS and 0.1% allelic fraction for mutation detection. Very sensitive assays are needed which can increase error rates for first generation assays.
- Only two 10 mL tubes of blood are typically drawn from the mother or from the cancer patient. We do not have the ability to collect 500 mL units of blood in order to manufacture panels for suitable comparison testing.
- Pre-analytical sample preparation protocols must be followed precisely and use special collection tubes to prevent peripheral blood cell genomic DNA from contaminating the sample.
- There is a lack of suitable reference materials for assay development, run QC, method comparisons and proficiency testing. The potential for errors will be high during initial introduction but should drop with further optimization.
- Multiple formats are used: massively parallel sequencing, arrays and SNPs for NIPS, and real-time qPCR (RT-qPCR), digital PCR (dPCR) and NGS for ctDNA. All assays have tradeoffs and clinicians must be aware of these challenges.
- The majority of assays are Laboratory-Developed Tests (LDTs) with proprietary bioinformatics pipelines. Assays may or may not have undergone the rigors and transparency of interlab analytical performance testing or proficiency testing.
- NIPS is 2-3 years ahead of ctDNA in commercial use thus has moved past the “peak of inflated expectations” (a term coined by the Gartner advisory firm, commonly known as part of the ‘Hype Cycle’1) and is entering a phase of “adoption” while ctDNA is just entering the “peak of inflated expectations” phase.
So are there lessons learned from NIPS that can be applied to ctDNA?
The first is to plan on full transparency at even the early stages of development. Last year there was an Editorial by Jani et al2. titled “Cell free DNA testing: How to choose which laboratory to use?” that stands out for me. For labs, taking the clinician’s or patient’s point of view is certainly enlightening on what to report. The authors concisely summarized the challenges facing OB GYN doctors when choosing labs for fetal genetic screening.
The author’s checklist, which is a “transparency” checklist, includes:
- Knowing the origin of the assay
- Knowing if the assay is an LDT or has been acquired
- Reviewing first-hand the analytical and clinical validation data
- Asking for method comparisons, concordance data, sensitivity, false positive and false negative rates, test quality data and failure rate data
In their Editorial, Jani et al. also reported NIPS laboratory failure rates from Gil et al3. that range from 0.03% to 11.1%. This is a high and wide range and should be very disconcerting for a clinician selecting an assay.
So what else can be learned from this study? Gil et al. performed meta-analysis of 37 NIPS published studies. After removing the sex chromosome aneuploidy results from the data, since these assays are not as widely available, there were 30 reports listing assay failure for high risk aneuploidies Trisomy 13 (T13) also known as Patau Syndrome, Trisomy 18 (T18) also known as Edwards Syndrome and Trisomy 21 (T21) also known as Downs Syndrome. For this data set, the median error rate was 3.0% and the range was 0.8%-8.8%. The distribution for total errors is shown in Figure 1.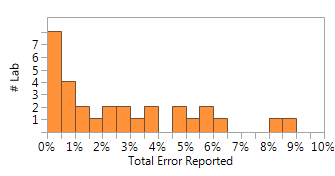
Figure 1. Histogram showing %Total Error by Labs (n=30) reporting. Gil et al.
Gil et al. also categorized the types of errors into two categories as Low Fetal Fraction or “Other” assay error for 9 laboratories. This breakdown is shown in Table 1.
| Type of Error | Median | Range |
| Low Fetal Fraction <4% | 1.80% | 0.5%-6.1% |
| Assay Error | 2.00% | 0.2%-2.8% |
Table 1. Breakout by Type of Error (n=9). Gil et al.
A 3% median error rate is high, even for a screening assay that requires confirmatory testing, and it points to significant due diligence challenges for clinicians. The assays surveyed by Gil et al. are first generation screening assays and as expected have trade-offs in performance. A high error rate is not unexpected since the technology is at an early stage. Reagents, instruments and software are minimally Research Use Only (RUO), there are no FDA-cleared assays to reference and there are few interlab studies or proficiency testing materials for these types of assays.
The advance of NIPS over the last 5 years and its rapid replacement of biochemical assays were done under yesteryears’ CLIA oversight. This will not be the case for ctDNA as the FDA is likely to impose stricter regulations directly on the CLIA laboratories <http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm407296.htm> or stricter regulations indirectly through CLIA modernization <http://www.mlo-online.com/Labline/201609/14/toc.htm>. It is guaranteed that increased visibility for greater awareness will be applied to ctDNA that was not applied early on to NIPS.
In summary, what lessons for the laboratory have been learned from NIPS testing that would help advance ctDNA assay development and help oncologists choose the right laboratory at the right time?
- Anticipate regulatory oversight and be very transparent early in assay development. Compared to NIPS, ctDNA assays require higher sensitivity (0.1%AF) and specificity so extensive analytical and performance testing should be reviewed before clinical trials are underway. Performance metrics should be established up front, even for analytical validation. Establish benchmarks by executing interlab studies using the same reference materials.
- Use manufactured reference materials or “contrived samples” that are designed and manufactured to challenge the assay. Use a wide range of allelic frequencies and highly multiplexed controls that contain most if not all variants. Use commutable like matrix.
There was a scarcity of reference materials during the early NIPS assay development stages. This is not the case for ctDNA as a number of ctDNA products are either available or in development today. We have several ctDNA reference material products manufactured using biomimetic technology to assist in this effort - click here for more details. http://www.seracare.com/oncology. SeraCare’s patent pending biomimetic technology is used to develop the Seraseq™ Aneuploidy products which are used primarily in the development and validation of NIPS and Seraseq™ liquid biopsy ctDNA products. Biomimetics are used to make patient-like circulating DNA reference materials for all testing phases of assay development through daily run QC. Long-term stability is another key advantage of biomimetic technology since DNA not prepared using biomimetic technology is unstable in plasma leading to erroneous results.
References:1. https://en.wikipedia.org/wiki/Hype_cycle
3. Gil, M. M., et al. (2015). "Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: updated meta-analysis." Ultrasound Obstet Gynecol 45(3): 249-266. https://www.ncbi.nlm.nih.gov/pubmed/25639627
OBJECTIVE: To review clinical validation or implementation studies of maternal blood cell-free (cf) DNA analysis and define the performance of screening for fetal trisomies 21, 18 and 13 and sex chromosome aneuploidies. METHODS: Searches of PubMed, EMBASE and The Cochrane Library were performed to identify all peer-reviewed articles on cfDNA testing in screening for aneuploidies between January 2011, when the first such study was published, and 4 January 2015. RESULTS: In total, 37 relevant studies were identified and these were used for the meta-analysis on the performance of cfDNA testing in screening for aneuploidies. These studies reported cfDNA results in relation to fetal karyotype from invasive testing or clinical outcome. Weighted pooled detection rates (DR) and false-positive rates (FPR) in singleton pregnancies were 99.2% (95% CI, 98.5-99.6%) and 0.09% (95% CI, 0.05-0.14%), respectively, for trisomy 21, 96.3% (95% CI, 94.3-97.9%) and 0.13% (95% CI, 0.07-0.20) for trisomy 18, 91.0% (95% CI, 85.0-95.6%) and 0.13% (95% CI, 0.05-0.26%) for trisomy 13, 90.3% (95% CI, 85.7-94.2%) and 0.23% (95% CI, 0.14-0.34%) for monosomy X and 93.0% (95% CI, 85.8-97.8%) and 0.14% (95% CI, 0.06-0.24%) for sex chromosome aneuploidies other than monosomy X. For twin pregnancies, the DR for trisomy 21 was 93.7% (95% CI, 83.6-99.2%) and the FPR was 0.23% (95% CI, 0.00-0.92%). CONCLUSION: Screening for trisomy 21 by analysis of cfDNA in maternal blood is superior to that of all other traditional methods of screening, with higher DR and lower FPR. The performance of screening for trisomies 18 and 13 and sex chromosome aneuploidies is considerably worse than that for trisomy 21.
2. Jani, J., et al. (2015). "Cell-free DNA testing: how to choose which laboratory to use?" Ultrasound Obstet Gynecol 46(5): 515-517. https://www.ncbi.nlm.nih.gov/pubmed/26300279




