SeraCare’s clinical genomics technologies are developed to address challenges faced across the spectrum of NGS assays. From early development of assays – either IVD assay manufacturers or clinical labs building their own LDTs - there is a scarcity of characterized, complex, difficult variants to ensure the assay can robustly detect all the critical genomic variants in a patient sample. Using our highly characterized, reproducible, and GMP-grade NGS standards, laboratories have a wide range of analytical and clinical validation tools to deeply characterize assay performance such as LOD, linearity, specificity, sensitivity, and reproducibility.
Once a lab has validated its assay, we provide tools for implementing a robust NGS QC system including precisely manufactured daily run controls as well as software to track all NGS QC metrics in the NGS workflow. Lastly, by having consistent and reproducible universal QC standards, labs are able to “talk” to one another despite their different assay formats or platforms and engage in external quality assessment and proficiency programs.At the recently concluded 2018 Next Generation Dx Summit, SeraCare presented data on its liquid biopsy reference standards, including a large multi-lab investigation to assess the use of reference materials in quality assurance determinations for liquid biopsy-based NGS assays. The multi-lab, blinded study was coordinated by scientists from the National Institute of Standards (NIST), and involved 6 laboratories and organizations – Johns Hopkins Medicine, Boston University Medicine, National Cancer Institute, UNC School of Medicine, Göteborgs Universitet, and SeraCare Life Sciences.
In evaluating assay sensitivity across different technologies and different laboratories, DNA mutations in the Seraseq ctDNA v2 mutation mix AF0.125% reference standard were analyzed by digital PCR and targeted NGS panels at two different sites (E & F); results are shown in Figure 1. Both platforms reliably detected all mutations assayed at 0.125% with good concordance across the different technologies. Details of the completed multi-lab investigation results will be published by the laboratory investigators.
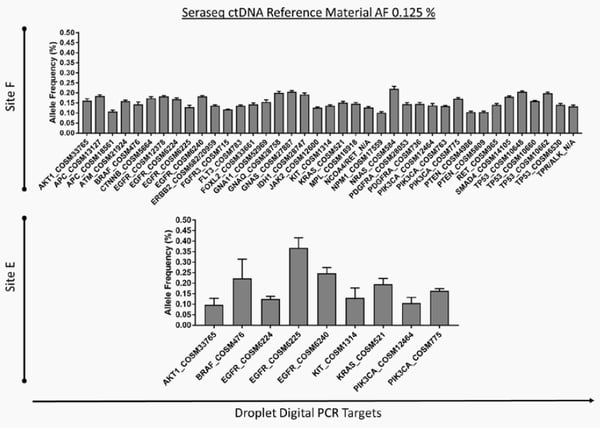
Figure 1: NGS (top) and digital PCR (bottom) analysis of DNA variants at 0.125% VAF in the Seraseq® ctDNA v2 mutation mix, from Lab F and Lab E, respectively [Data from the 2018 AACR Poster#3657, by E.V. Stein, et. al.]
The Seraseq ctDNA reference materials have found wide applications in the development and validation of NGS assays, supporting AF sensitivity claims, as well as assay robustness. Table 1 highlights some current cfDNA-based NGS and multiplex PCR assays that have been evaluated using Seraseq ctDNA reference standards.
| Vendor | Panel | # of genes | AF% range | Platform |
| Archer | Reveal ctDNA 28 | 28 | 1-5% (or as low as 0.25%) | MiSeq/NextSeq |
| Paragon | CleanPlex® OncoZoom® Panel | 65 | 1% | MiSeq/NextSeq |
| Swift | Accel-Amplicon™ Panels (56G) | 56 | 1% | MiSeq/NextSeq |
| Roche | AVENIO Targeted | 17 | 0.5-1% | NextSeq |
| Agena | UltraSeek Lung | 5 | as low as 0.1% | MassArray |
Table 1: Examples of ccfDNA-based assays evaluated with Seraseq ctDNA reference standards.
In conclusion, these studies highlight the application of Seraseq ctDNA reference materials in supporting the development, validation, and clinical implementation of ccfDNA-based assays (NGS or multiplex PCR), as well as providing a ready source of critical patient-like reference samples for liquid biopsy-based clinical patient diagnostic testing and disease management.
For more details about SeraCare’s ctDNA reference samples please visit our liquid biopsy portfolio.
Free Poster and Video Download:
Multi-Laboratory Assessment of a New Reference Material for Quality Assurance of Circulating Tumor DNA Measurements
 Download the poster and watch this video to see how the development of manufactured reference materials in large consistent quantities enable testing labs to compare results and check quality and traceability at extraction, library prep, sequencing, and analysis.
Download the poster and watch this video to see how the development of manufactured reference materials in large consistent quantities enable testing labs to compare results and check quality and traceability at extraction, library prep, sequencing, and analysis.





