An important goal in cancer disease management is early detection. When detected early, disease progression can be significantly mitigated with a plethora of options (targeted therapy, chemotherapy, surgery, etc.) available to medical practitioners, to afford progression free survival and a higher quality of life. A great promise of liquid biopsies is the possibility of early detection of cancer long before clear evidence of lesions and tumor growth observable by imaging or other techniques.1 As proxy for solid tissue biopsies, plasma-based liquid biopsy application is rapidly gaining traction in cancer disease diagnosis, progression, monitoring, and in predicting resistance to treatment options.2
Next-generation sequencing (NGS) is the leading clinical genomics platform for developing and deploying assays that use circulating cell-free DNA (ccfDNA) samples in cancer patient testing. In order to expedite the clinical implementation and standardization of ccfDNA-based NGS assays, SeraCare has developed a portfolio of circulating tumor DNA (ctDNA) reference materials that mimic cancer patient plasma profiles (ccfDNA sizing), are highly multiplexed (25-40 variants per sample), allow for the broadest analysis of clinically relevant variant types (SNVs, INDELs, CNVs, and SVs), and are engineered with VAFs as low as 0.1% to address every phase of the assay process from development to the clinic.
These products, in purified ctDNA or plasma-like encapsulated formats, allow end-to-end sample workflow process control for liquid biopsy-based NGS assays. Clinical NGS laboratories can now process plasma reference samples alongside patient plasma samples, from sample extraction to clinical data report generation, while incorporating ground truth highly-multiplexed variants in ctDNA reference materials.
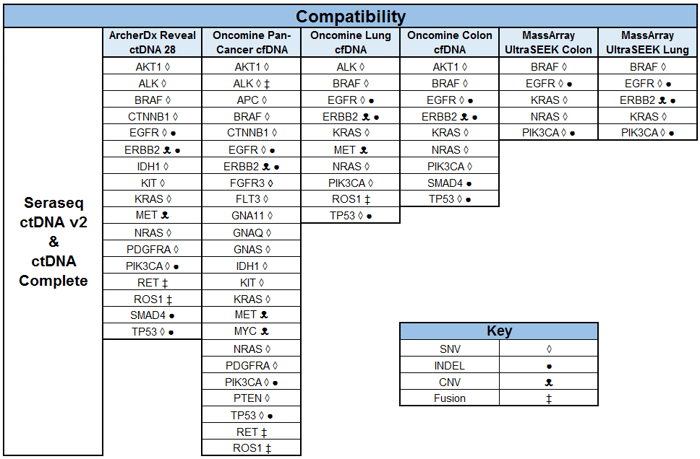
The Seraseq® ctDNA gene content was developed in consultation with a consortium of industry experts in molecular pathology, clinical oncology and cancer biology, as well as published information on clinically-guided biomarkers (NCCN, ASCO, ESMO, AMP) with on-market or in-clinical trial therapeutic agents. The below table provides a summary of Seraseq ctDNA reference material compatibility (ctDNA v2 and ctDNA Complete™) with several on-market ccfDNA targeted NGS assays3 including ArcherDx® Reveal ctDNA™ 28, Oncomine™ Pan-Cancer cfDNA, Oncomine™ Lung cfDNA, Oncomine™ Colon cfDNA, MassArray™ UltraSEEK Colon, and MassArray™ UltraSEEK Lung.
To demonstrate the use of ctDNA reference samples in enhancing the efficiency of ccfDNA-based NGS analysis of cancer patient samples, a study of plasma samples from normal and diseased patients, as well as reference samples from vendors such as SeraCare and others (see Figure 1) was conducted.4 In the study, cell-free DNA from mixed cell lines from a commercial vendor,5 SeraCare,6 and cancer patient plasma samples, healthy controls as well as mutation-positive patient plasma admixtures, were uniquely tagged with a random molecular barcode (MBC), amplified in an efficient PCR protocol, and sequenced using a targeted panel covering >500 somatic mutation hotspots. The molecular diversity of the barcoded samples (MBI) was determined and plotted for all amplicons in all samples. Samples with median MBIs greater than 100% of expectation were associated with a highly efficient sequencing analysis and the potential for detection of low frequency ctDNA variants at 0.1% VAF.
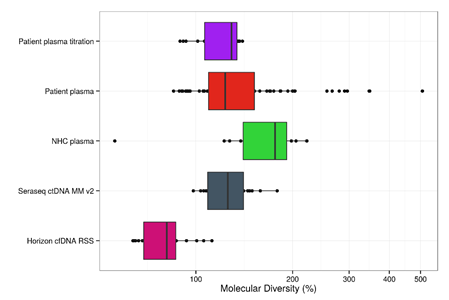
Figure 1: Post-sequencing analysis of molecular diversity of barcoded molecules plotted for all amplicons in sequenced samples from SeraCare, mixed cell line-based, normal and diseased patients [JL Larson, et. al., 2018 AACR Poster]. % Molecular Diversity is the # of observed read groups post-sequencing divided by the number of template molecules (DNA haploid genome equivalents estimated by Qubit using a standard conversion factor).
In conclusion, the development of these new generation of highly multiplexed and “patient-like” circulating tumor DNA reference samples meet today’s need for sourcing requisite samples with the breadth of genomic variants and sensitivity required for the development and validation of highly sensitive liquid biopsy assays. These new highly multiplexed ctDNA reference materials allow clinical laboratories and NGS assay vendors to accelerate the development, validation, and clinical deployment of liquid biopsy-based NGS testing, advancing the promise that precision and personalized medicine has long offered.
For more details about SeraCare’s ctDNA reference samples please visit our liquid biopsy portfolio.
1 S. Perakis & M.R. Speicher; Emerging Concepts in Liquid Biopsies, BMC Medicine, 2017, 15: 75.
2 E.A. Ashley; Towards Precision Medicine, Nat. Rev. Genet., 2016, 17(9): 507.
3 Circulating cell-free DNA NGS targeted sequencing panels from ArcherDx® (Reveal ctDNA® 28), Thermo Fisher (Oncomine™), and Agena Biosciences (MassArray®).
4 Poster #5574 / 6 – 2018 American Association of Cancer Research (AACR): A comprehensive, targeted next-generation sequencing method that rapidly and accurately detects circulating tumor DNA variants at 0.1% frequency in plasma samples – JL Larson, L Chien, L Ringel, B Printy, FL Tomson, Y Konigshofer, S Statt, JK Kaplan, S Gokul, J Shelton, GJ Latham, BC Haynes.
5 https://www.horizondiscovery.com/reference-standards/our-formats/cell-free-dna
6 https://www.seracare.com/en-US/about-us/customers/precision-medicine/liquid-biopsy-standards/
Are you looking for clarity on validation guidance?

Our new eBook offers insights for more effective and less costly assay validation and will walk you through key considerations and guidelines for success.
"Nothing is more frustrating than finding a sample that is positive for a relevant variant but cannot be tested multiple times due to sample depletion."





