Clinical genomics laboratories are increasingly looking to liquid biopsy cancer assays to complement their current solid tumor assays. Compared to their solid tumor assay counterparts, circulating tumor DNA (ctDNA) assays offer a different set of challenges to consider for clinical labs. One of the most important of which, is to develop a set of reagents that are appropriately validated to determine the critical performance of the assay across many parameters. The ctDNA targets of liquid biopsy assays are typically at much lower allelic frequencies and require a robust and reproducibly designed assay to consistently detect these important variants.
Few good options available to labs
Even if you are fortunate to have access to a large patient specimen repository, developing and validating even the most simple of liquid biopsy tests will quickly drain these resources. Also the ready availability of specimens with clinically important but rarer DNA variants is often limited.
It is important that all of these specimens have been evaluated by a separate validated assay to confirm presence of target variants at a specified allelic frequency before being used to validate a new assay.
It is no wonder that to do a properly designed development and validation program—contrived and commercially derived reference materials will need to play a major role for labs. The trouble is that current options have been significantly lacking in the ability to emulate the performance of native cell-free DNA (cfDNA) which is critically important to extrapolate these performance data sets to those of real patient specimens.
Three problems with cfDNA reference materials
- DNA size
The size of the DNA in the reference material is very important to its performance. cfDNA has a narrow size profile—on average around 170bp in size, and assays need to be optimized to detect targets in this size range. Reference materials should exhibit a similar size distribution to native cfDNA in order to work similarly in the assay.
- Stability and function
A commonly used method has been to use genomic DNA from cancer cell lines, either diluted in normal plasma or first sheared using sonication and then diluted. While one can target an average fragment size of ~170bp with sonication, the resulting mix often has a very wide distribution in size, often much more broad than profiles for native cfDNA. These broader populations of target DNA do not perform nearly as well as native cfDNA with much of this DNA not amplifiable, and thus not able to be incorporated into NGS libraries. This has important ramifications for library yield and genomic representation for labs to overcome.
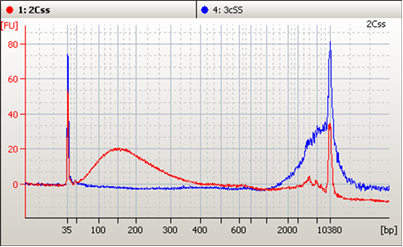 CfDNA from human plasma isn’t simply DNA in plasma. While natural cfDNA has a short half-life it is still associated with lipids and proteins and other molecules in the matrix. Simply mixing fragmented, purified DNA into normal plasma does not result in a stable mixture. Figure 1 shows a size trace comparing stabilized and non-stabilized fragmented DNA in a synthetic human plasma (SeraCon™ Matribase Negative Diluent), with a noticeable aggregate >3,000 bp and a simultaneous dramatic decline in the amount of 170 bp fragments. The result is that very quickly, DNA in the non-stabilized form is no longer available for detection. So for labs wanting to evaluate their full assay workflow, reference materials that behave more like real patient samples are required.
CfDNA from human plasma isn’t simply DNA in plasma. While natural cfDNA has a short half-life it is still associated with lipids and proteins and other molecules in the matrix. Simply mixing fragmented, purified DNA into normal plasma does not result in a stable mixture. Figure 1 shows a size trace comparing stabilized and non-stabilized fragmented DNA in a synthetic human plasma (SeraCon™ Matribase Negative Diluent), with a noticeable aggregate >3,000 bp and a simultaneous dramatic decline in the amount of 170 bp fragments. The result is that very quickly, DNA in the non-stabilized form is no longer available for detection. So for labs wanting to evaluate their full assay workflow, reference materials that behave more like real patient samples are required.
Figure 1: Size-based Agilent BioAnalyzer 2100 analysis of 170 base-pair (bp) fragmented DNA blended with SeraCon Matribase with (red) and without (blue) stabilization after 66 days at 42 °C. Note the presence of aggregated product >3,000 bp in blue.
3. Mutation coverage
The number of clinically important variants and variant types continues to grow with ongoing clinical research into the utility of liquid biopsy assays. For labs to have the utmost confidence in their assay’s ability to detect the widest possible range of clinically important variants—they need these specimens for their validation. DNA from cell lines can play an important role here, as would characterized patient specimens. However, there is typically always gaps in coverage for all the variants. Cost is a very big factor as well. Having access to specimens (cell lines or other) with these variants is typically 1 variant per specimen. It proves time consuming and costly for labs to sequentially test these specimens, and as a consequence tradeoffs are typically made between number of specimens and the overall testing costs.
We have a solution
SeraCare has solved these three problems with a new set of patient-like ctDNA reference materials developed with patent-pending technology. The Seraseq™ ctDNA v2 reference materials offer laboratorians size appropriate DNA materials (Figure 2) that function similarly to native cfDNA specimens in clinical genomics assays. These materials are available in purified and synthetic plasma formats, offer the broadest collection of important ctDNA variants in a single tube (Table 1), and include precisely quantitated allelic frequencies from 2% to 0.125%.
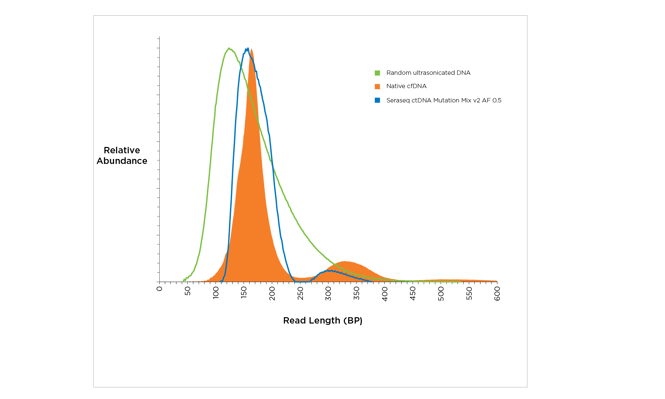
Figure 2: Three types of fragmented DNA: random ultrasonicated (green), native cfDNA (orange), and the Seraseq™ ctDNA Mutation Mix v2 AF 0.5 (blue). Samples were characterized by the Agilent 2100 Bioanalyzer®.).
|
AKT1 |
APC |
BRAF |
CTNNB1 |
EGFR |
ERBB2 |
|
FGFR3 |
GNA11 |
GNAQ |
GNAS |
IDH1 |
JAK2 |
|
KIT |
KRAS |
MPL |
NPM1 |
PDGFRA |
PIK3CA |
|
PTEN |
RET |
SMAD4 |
TP53 |
NRAS |
Table 1: List of 23 actionable genes in the Seraseq Circulating DNA V2 products. See the full list of variants here.
Learn more about our Seraseq ctDNA V2 products or download our technical report to see data and in-depth performance information.





