One of several important steps in next-generation sequencing (NGS) is tuning the many options provided by mutation callers. Providing values for options configures the signal to noise ratio of the impending mutation calls. In theory, providing values that increase the stringency of mutation calls will reduce the number of false positive calls and thus enrich for true positives. In practice, increasing stringency can eliminate true positives.
We noticed this phenomenon while QC-testing our second lot of Seraseq ctDNAv2 Mutation Mix AF1%. The allele frequency of one of the mutations, COSM763 (PIK3CA p.E545K ), dropped substantially when testing the second lot as compared to the first lot (AF=0.12% versus AF=0.82%, respectively) in spite of using the same amplicon assay and sequencing platform. Quizzically, the other mutations contained in the mutation mix did not demonstrate proportional changes with respect to COSM763. Upon inspection of the sequence alignments with the Integrated Genome Viewer (IGV), we found that 20 of the total 1821 reads (AF=1.10%) contained the mutant A allele (Figure 1).
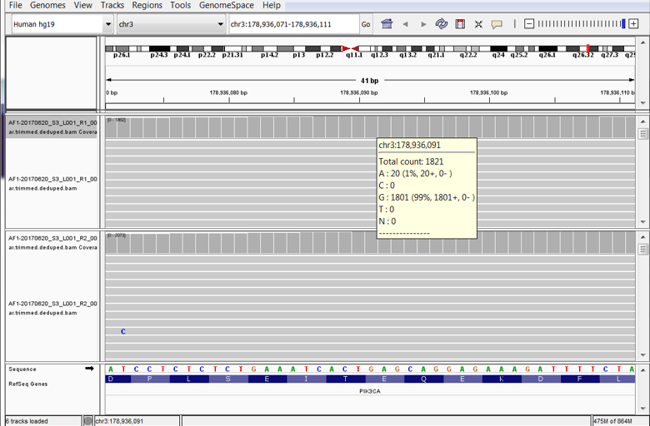
Figure 1. IGV screen shot of the PIK3CA region. A pop-up of the base coverage at the COSM763 position (yellow box) demonstrates that 20 of the total 1821 mutations are the expected A mutant allele conforming well with the expected allele frequency of 1.0%.
If the COSM736 mutations are present in the sequencing reads, why are they not being called by the mutation caller? Further interrogation of the sequence alignments in IGV revealed the answer. The majority of mutant sequence reads had MAPQ scores below 30 (Figure 2). The MAPQ score is a phred value that quantifies whether a sequencing read has been incorrectly placed in the reference genome. One might ask, why would the MAPQ scores change from Lot 1 to Lot 2. The simple answer is that the MAPQ scores did not change, the options provided to mutation caller in our pipeline had been updated. We had increased the MAPQ minimum threshold option from 20 to 30. This alteration equates to raising the accuracy of placement in the reference genome from 99% to 99.9%. The increased threshold caused mutations that were originally called for Lot 1 not to be called for Lot 2.
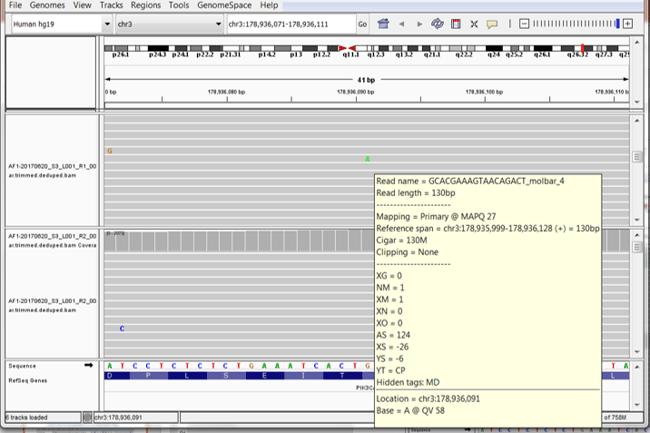
Figure 2. Screen shot from IGV with a pop-out (yellow) showing a sequence read containing the COSM763 (A allele). The MAPQ value of this read is 27 (red arrow) which falls below the minimum MAPQ threshold setting of 30.
This example nicely demonstrates some important points regarding NGS assays and informatics. The first is that not all parts of the genome are created equal. This is not a surprising revelation but may be underappreciated. As compared to the other mutations in our mix, the single base change of COSM763 was enough to cause most mutant reads to have MAPQ values in the low to mid 20s. By defining a MAPQ minimum threshold value of 30, this true positive mutation call was missed.
In addition to the non-uniformity of the human genome, this COSM763 drop out example exhibits the need for NGS assay developers to test prevalent and clinically relevant mutations directly. Just because you can detect some mutations in your gene of interest does not mean that you can detect ALL mutations in your gene of interest. Therefore, if there are recurrent mutations that cause your disease of interest, reference controls should be used to evaluate the ability of your assay to detect and correctly call those mutations. In this regard, SeraCare’s VariantFlex Custom NGS Library product is ideal for this purpose. VariantFlex enables one to calibrate the informatic options of a mutation caller to a spectrum of mutations of their choosing.

Free eBook: Next-Generation Sequencing Assay Validation
A Practical Guide for the Clinical Genomics Laboratory
This eBook focuses on the validation process for NGS-based assays and will walk through key considerations and guidelines you can follow to ensure a smooth and successful validation.
Next-generation sequencing (NGS) has revolutionized the field of genomics and how in vitro diagnostic (IVD) test developers, laboratories, and clinicians are diagnosing, treating, and monitoring disease. Before you can successfully launch a clinical assay, platform, or service, you need to be absolutely confident that the test has gone through rigorous development and thorough validation to ensure accuracy of the result.





