Authors: Yves Konigshofer, PhD; Andrew Anfora, PhD; Omo Clement, PhD; and Krystyna Nahlik, PhD. LGC Clinical Diagnostics.
Introduction
Liquid biopsy methods developed for circulating tumor DNA (ctDNA) analysis in solid tumors are transforming clinical practice, allowing for non-invasive detection and assessment of earlier stages of disease, monitoring for resistance to therapy, and post-treatment monitoring for minimal residual disease (MRD) and recurrence of cancer. The presence of minimal residual disease may be prognostic in that is has been found to correlate with worse patient outcomes, so early and accurate measurement is crucial. ctDNA-based assays allow for the detection of MRD at earlier time points than standard clinical and imaging surveillance, and could allow for treatment modification based on real-time assessment of the tumor genomic landscape.
Many challenges remain, as the analytical validation of liquid biopsy-based MRD assays requires reference materials allowing for the assessment of sensitivity and specificity at variant allele frequencies (VAFs) that can be over an order of magnitude below the typical limit of detection of standard ctDNA assays 1 . Current MRD testing is much more tied to NGS than are other personalized medicine approaches, where it may be practical to interrogate specific mutations via PCR-based methods. At such low VAFs, there may be on average less than one copy of a somatic variant in a sample being analyzed out of billions of ctDNA fragments in a blood draw. This “fraction of a needle in a haystack” problem is why many MRD approaches use whole exome sequencing data from patient tumors to design tumor-informed, personalized assays that target multiple patient-specific somatic variants, which reduces the amount of sequencing required to survey MRD while allowing the sequencing depth around the targeted variants to reach a level that is sufficient for high sensitivity and specificity. Even after reducing the variant list to a handful of patient-specific molecular signatures, robust surveillance of MRD requires the high coverage at low cost that only NGS can currently provide 2 . The urgent need for suitable reference materials for the validation of such NGS assays is highlighted by the recent application of serial dilutionsas applied in the Natera Signatera3 and TGEN’s Targeted Digital Sequencing (TARDIS)4 MRD assay validation.
Creation of a novel MRD reference material
LGC SeraCare has developed a novel ctDNA MRD panel designed to support the development, validation and clinical deployment of ctDNA-based MRD monitoring NGS assays. The Seraseq ctDNA MRD Panel Mix product is derived from a mixture of tumor human cell line gDNA, its SNP-matched normal gDNA and biosynthetic DNA containing 22 additional variants commonly analyzed in targeted NGS assays. The DNA was fragmented, sized to mimic ccfDNA, and serially diluted to obtain tumor fraction (TF) blends of 0%, 0.5%, 0.05% and 0.005%. The synthetic variants were blended at 0.25% target VAF for the 0.5% TF blend to represent an average 50% allele frequency in the tumor.
After fragmentation, 50 ng of DNA was used as input for a custom Agilent SureSelect XT HS2 assay that targets 100 somatic variants from over 600 identified by WES in the tumor cell line, which is sufficient to confirm if the genome of the cell line is represented as expected in all finished tumor fraction blend levels as shown in Figure 1.
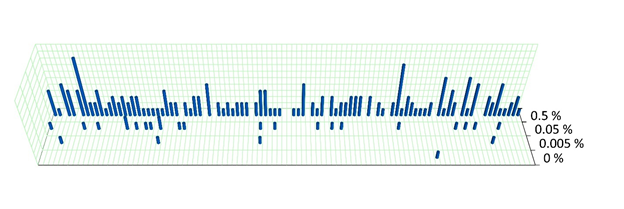
Figure 1. Visual representation of unique variant observations detected with the 100 somatic variant target panel at each TF. The vertical axis shows the number of observations of the targeted variants after error correction.
Somatic variants in the 0.5 % and 0.05 % samples were detected as expected (see Table 1), while the 0.005 % and 0% sample had 4 and 1 variant detected, respectively. However, the number of observed variants generally conforms to the expected number for the given tumor content shown in Table 1.
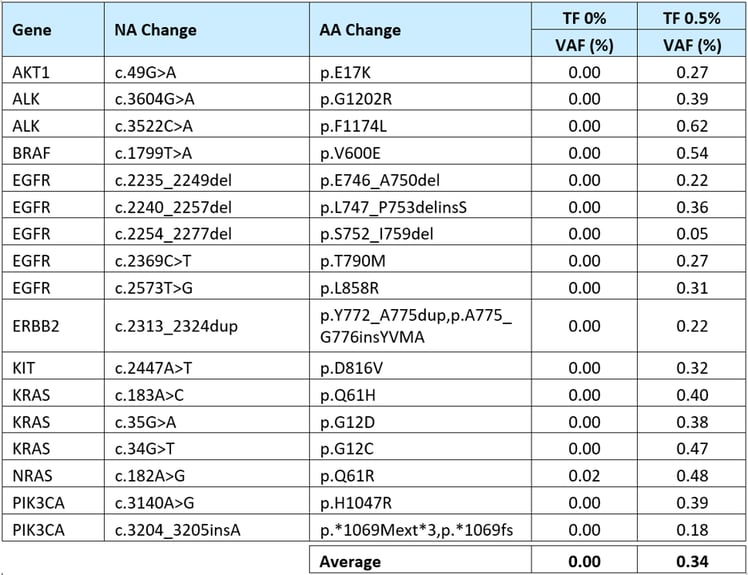
Table 1. Estimated number* of detectable variants (out of a subset of 100) for TFs at 0.5%, 0.05%, and 0.005 % MRD materials (low and high are the bounds of the 95 % CI).

*Estimate is based on the number of analyzable molecules and an expectation of average VAFs around 50 % of the tumor content.
The majority of variants are coded in the genome of the tumor cell line and are present at a range of VAFs between ~0 to 1% in the 0.5% tumor blend due to the aneuploidy of the tumor cell line. Fragmentation, ploidy, and depth of sequencing have minor impact on the observed AF of expected variants, but the material generally performs as expected for cfDNA at targeted TF.
Verification of correct dilution to low VAF
As expected, increasingly fewer variants were detected at increased dilutions of tumor fraction. There is currently no set standard for MRD data analysis best practices and the expectation is that the majority of mutations are present in all of the MRD TF preparations if high enough amounts of the samples were sequenced at sufficiently high levels of coverage depth. It should also be noted that while doubling the DNA input mass would increase the likelihood of including very low frequency variants, it would also lead to more background noise and a challenge to adequately error-correct. Thus, to improve sensitivity without reducing specificity, doubling the DNA input mass would likely require more than doubling the amount of sequencing per sample.
Table 2. Aggregate summary of cell line-derived variants detected at each tumor fraction (TF) in the MRD reference material.
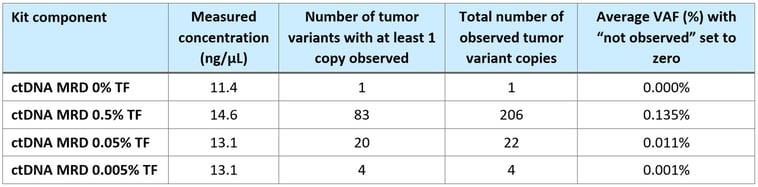
To confirm the presence of the synthetic targetable variants, the TF 0% and 0.5% MRD reference materials was analyzed by ArcherDX Reveal ctDNA-28 kit which can detect 17 of the 22 spiked-in synthetic variants (concordant content). MRD samples at 0.05% and 0.005% TFs were not tested because of the NGS assay’s limited sensitivity/specificity at VAF <0.25%.
As shown in Table 3, the observed VAFs for the spiked-in 17 variants were generally consistent with the 0.5% TF and 0% TF after error correction.
Table 3: Measured VAFs for the biosynthetic variants spiked into the ctDNA MRD Panel Mix.
Confirmation of cfDNA-like fragment profile
Much like other Seraseq liquid biopsy products, the MRD material represents a patient-like ctDNA fragment size. The expected somatic cell line variants show a similar fragment size distribution as the remainder of the genome, see Figure 2.
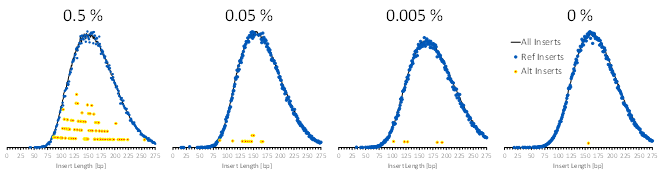
Figure 2. Agilent XT HS2 analysis of the somatic MRD variants after error correction for the insert size distribution of MRD samples. Insert sizes are shown for all reads (black lines), reference sequence-containing reads (blue dots), and variant-containing reads (red/yellow dots).
The fragments derived from synthetic DNA are similar in length to the inserts derived from the genomic cell line backbone gDNA in Archer Reveal ctDNA 28 libraries as shown in Figure 3. The data of the variant-containing reads is noisier due to the significantly lower abundance of such reads. It should be noted that because of the nature of this assay, the inserts do not reproduce the fragment lengths of the input DNA but rather a convolution thereof.
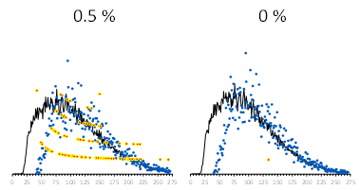
Figure 3. Insert size distribution for all reads (black lines), reference sequence-containing reads (blue dots), and variant-containing reads (red/yellow dots). ArcherDX Reveal ctDNA-28 of the synthetic variants.
Conclusion
We have created a reference material combining a tumor-derived component with a sufficiently large number of somatic mutations to accommodate the design of custom tumor-informed MRD assays2,5. Additionally, the 22 synthetic variants can be used in the evaluation of fixed ctDNA panels. The reference material contains DNA fragments with a similar size distribution to that of cfDNA isolated from patient samples. It has been supplemented with variants representing frequent solid tumor driver ad resistance mutations which are likely to be included in both general and personalized ctDNA panels. This well-characterized and unbiased reference material is a unique tool to support the development and validation of MRD assays based on the detection of patient-specific variants for MRD monitoring at very low allelic frequencies.
References:
- Yves Konigshofer, MRD blog post, part 1
- Yves Konigshofer, MRD blog post, part 2
- Murtaza, M. et al., Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer Transl. Med. 11, eaax7392 (2019)
- Magbanua MJM et al., Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival, Annals of Oncology (2020), doi:10.1016/j.annonc.2020.11.007.
- Yves Konigshofer, et. al., “Reference Materials for Measurable Residual Disease (MRD) Monitoring in Circulating Cell Free DNA (ccfDNA)” MRD Poster @ AMP 2020




