Choose your Article Focus | NGS | Molecular & Serology
Yves Konigshofer, PhD
Recent Posts
Development of a Novel Reference Material for MRD Monitoring Assays
Category: MRD, ccfDNA, cfDNA, Minimal Residual Disease, NGS, ctDNA, reference materials
Posted by
Yves Konigshofer, PhD on Aug 17, 2021 12:00:00 AM
Authors: Yves Konigshofer, PhD; Andrew Anfora, PhD; Omo Clement, PhD; and Krystyna Nahlik, PhD. LGC Clinical Diagnostics. Introduction Liquid biopsy methods developed for circulating tumor DNA (ctDNA) analysis in solid tumors are transforming clinical practice, allowing for non-invasive detection and assessment of earlier stages of disease, monitoring for resistance to therapy, and post-treatment monitoring for minimal residual disease (MRD) and recurrence of cancer. The presence of minimal residual disease may be prognostic in that is has been found to correlate with worse patient outcomes, so early and accurate measurement is crucial. ctDNA-based assays allow for the detection of MRD at earlier time points than standard clinical and imaging surveillance, and could allow for treatment modification based on real-time assessment of the tumor genomic landscape.
0 Comments Click here to read/write comments
How to resolve the challenges of MRD?
Category: MRD, cfDNA, NGS, ctDNA, reference materials
Posted by
Yves Konigshofer, PhD on Oct 7, 2020 12:00:00 AM
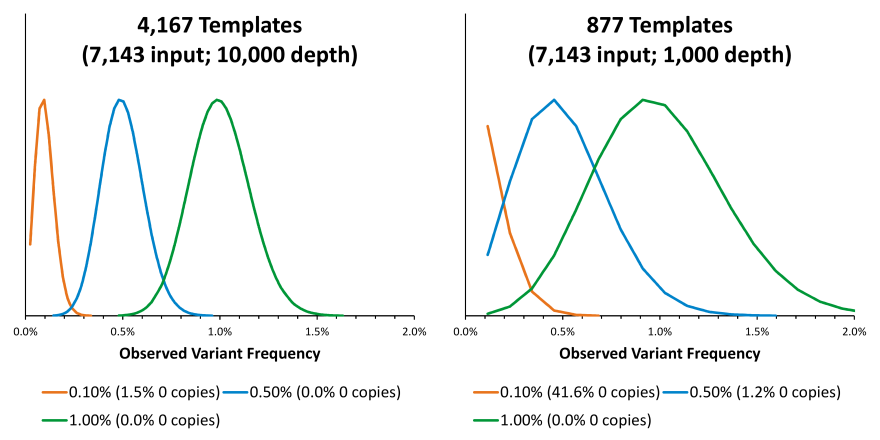
This is part 2 of 2 of the MRD blog post. (Click here for part 1). In this section, we will discuss how to overcome some of the most common challenges of MRD testing. Overcoming the Challenges In order to mitigate sequencing errors, methods using Molecular Barcodes (MBCs), Unique Molecular Identifiers (UMIs), etc. (which are essentially all the same) may be used, where each starting molecule is sequenced many times. The MBCs are then used to generate consensus sequences from sequences that were likely obtained from the same starting molecule. The assumption is that errors appear due to somewhat stochastic processes and that the consensus sequences will likely be correct. This requires many observations of the same starting molecule, so it will be recommended to generate 10-fold more sequences than there are molecules. Therefore, with 8,000 genomic equivalents, we might want to target a sequencing depth of 80,000. This is a reason why using 10-fold more input ccfDNA may not necessarily be a good thing (in addition to having to obtain a 10-fold larger liquid biopsy) since we may have to increase sequencing depth accordingly to 800,000, which could increase the cost of sequencing 10-fold, which could reduce the likelihood for payment and running the assay profitably.
0 Comments Click here to read/write comments
So, you want to monitor Measurable Residual Disease? What are the challenges?
Category: MRD, cfDNA, Minimal Residual Disease, NGS, ctDNA, reference materials
Posted by
Yves Konigshofer, PhD on Oct 1, 2020 12:00:00 AM
Part 1 of 2 Background Measurable Residual Disease (MRD) monitoring – for purposes of this blog – will be the act of looking for somatic variants in a liquid biopsy sample by analyzing circulating cell-free DNA (ccfDNA). This is done to monitor the disappearance of a metastatic solid tumor during treatment and to follow any future reemergence of that cancer. Analyzing ccfDNA assumes that circulating tumor DNA (ctDNA) will be present, and the median ctDNA frequency in patient samples across cancers seems to be around 0.5 to 1 %. Thus, the median variant allele frequencies (VAFs) of the somatic variants will start around this range, and the goal of MRD monitoring is to be able to detect them at much lower VAFs. This can be challenging and if we are going to design an assay for MRD monitoring, then we need to be aware of them and overcome them.
0 Comments Click here to read/write comments
The Full Authority Companion Diagnostic
Category: FDA, clinical genomics, NGS
Posted by
Yves Konigshofer, PhD on Feb 21, 2019 12:00:00 AM
It is very likely that on your last flight the turbofan engines were controlled by full authority digital engine controls – FADECs for short. FADECs have played a significant role in keeping airline ticket prices low (except during holidays) by continually adjusting engine parameters so that the engine operates with maximum fuel efficiency and within operational limits, allowing pilots to focus on other tasks.
0 Comments Click here to read/write comments
Developing a Rock-Solid Lung Cancer Assay, Part 2
Category: NGS, cancer, Assay Development, Lung Cancer
Posted by
Yves Konigshofer, PhD on Jul 2, 2018 12:00:00 AM
In a recent post, we discussed key considerations for designing a robust next-generation sequencing (NGS)-based lung cancer assay. Putting those plans into action in the development phase brings forth a new set of challenges. Through our experience developing NGS reference materials and the relationships we’ve built with assay developers of all stripes, we’ve identified those important factors and ways to navigate them. But before you begin designing and optimizing your assay, you should become very familiar with binomial and Poisson distributions and their use because the outcome of many analytical steps can be modeled and explained with them.
0 Comments Click here to read/write comments
Developing a Rock-Solid Lung Cancer Assay
Category: NGS, cancer, Lung Cancer, reference materials
Posted by
Yves Konigshofer, PhD on Mar 15, 2018 12:00:00 AM
Next-generation sequencing (NGS) allows deeper insights than ever before into the human genome and a host of diseases and conditions. So it makes sense that there is a worldwide movement to employ NGS in a growing number of applications. But as the saying goes, with great power comes great responsibility.
0 Comments Click here to read/write comments
Extracting Formalin-fixed, paraffin-embedded (FFPE) nucleic acids for NGS
Category: NGS
Posted by
Yves Konigshofer, PhD on Mar 14, 2017 12:00:00 AM
In the course of patient care, formalin-fixation and paraffin-embedding (FFPE) of biopsy tissue samples are routinely performed, where these samples can be analyzed by histology and archived to link the sample with clinical long-term follow-up. With the development of advanced NGS-based oncology gene panels, it is becoming increasingly important to consider pre-analytic variables when extracting nucleic acids from FFPE-treated samples. This post covers frequently asked questions (FAQs) around the extraction of nucleic acids from FFPE samples for downstream NGS analysis.
0 Comments Click here to read/write comments
FDA-AACR Liquid Biopsies in Oncology Drug and Device Workshop
Category: FDA, clinical genomics, NGS, cancer, LDT, ctDNA, reference materials
Posted by
Yves Konigshofer, PhD on Aug 8, 2016 12:00:00 AM
The presentations during the FDA-AACR Liquid Biopsies in Oncology Drug and Device Development Workshop on July 19, 2016 included several important pieces of information that will likely guide the development of assays and their review by the FDA.
0 Comments Click here to read/write comments
The FDA NGS-based Oncology Panels Workshop
Posted by
Yves Konigshofer, PhD on Apr 1, 2016 12:00:00 AM
The U.S. Food and Drug Administration on February 25, 2016 held an all-day public workshop entitled “Next Generation Sequencing-based Oncology Panels” at their White Oak campus in Silver Spring, Maryland. Companion diagnostics were a recurring theme of the initial FDA presentations at this Next Generation Sequencing-Based Oncology Panels Workshop and these important tests will be our focus here. The slides that were presented at the workshop are available (PDF) and are referenced below, and those presented by SeraCare’s CSO Russell Garlick start on page 196.
0 Comments Click here to read/write comments
Does synthetic DNA sequence behave the same as cell-line DNA?
Posted by
Yves Konigshofer, PhD on Feb 5, 2016 12:00:00 AM
A recurring question SeraCare has been asked is whether synthetic DNA sequences perform similarly to cell line-derived sequences. For example, it could be hypothesized that secondary structure and/or the GC content of the DNA around a particular amplicon might lead to a detection bias when only 400 bases on either side of the amplicon are present compared to when many kilobases are present.
0 Comments Click here to read/write comments