Choose your Article Focus | NGS | Molecular & Serology
As to Myeloid Diseases, Make a Habit of Three Things
Category: myeloid cancer reference materials, NGS
Posted by
Ram Santhanam on Dec 13, 2017 12:00:00 AM
Previously, we wrote about the unique capabilities that next-generation sequencing (NGS) offers the oncology clinic. NGS could mark the beginning of a shift away from “single-site” technologies such as FISH and PCR-based testing, in favor of comprehensive screening across many different targets at once.
0 Comments Click here to read/write comments
3 Steps for Building a Bulletproof Clinical NGS Assay: Step 3
Category: qc management, NGS, RNA fusion
Posted by
Russell Garlick, PhD on Dec 8, 2017 12:00:00 AM
What does it mean for an NGS assay to be bulletproof and why does your lab need it? In two previous blog articles (parts one and two), we’ve talked about the factors that go into making NGS assays that doctors can rely on to deliver targeted, lifesaving therapies to their patients. Bulletproof assays are the tests that make your lab a trusted name in the NGS field, a leader in a rapidly-growing market. But, as we’ve written, genetic sequencing is complex, expensive, and time-consuming. Therefore, finding ways to do it more efficiently, while maintaining the quality of your tests, is in the best interests of your lab and its customers. As a refresher, here are the three steps for building a bulletproof clinical NGS assay: Consulting with experts Outlining your validation and quality control (QC) strategies together Evaluating reference material options We’ve already covered the first two steps. In this article, we’ll look at the third one. Choosing the right reference material technology can help control the high validation and running costs of highly multiplexed assays.
0 Comments Click here to read/write comments
The First Comprehensive Myeloid Cancer Reference Materials for NGS Assays
Category: myeloid cancer reference materials, NGS, reference materials
Posted by
Ram Santhanam on Nov 8, 2017 12:00:00 AM
Myeloid cancers are “liquid” tumors that arise from the blood and bone marrow. These diseases have undergone greater study and characterization than perhaps any other type of cancer, largely due to the ease of accessing these cancer cells via a blood draw rather than a tissue biopsy, as for solid tumors. There are many different types and subtypes of these malignancies that are known to be caused by mutations in genes that encode proteins involved in cell signaling, transcription, epigenetic regulation, and splicing1. Before next-generation sequencing became available in the hematology/oncology clinic, high-resolution genetic analysis of myeloid cancers relied primarily upon site-specific methods such as Fluorescence in Situ Hybridization (FISH) and PCR-based assays. And, while other methods such as karyotyping and array comparative genomic hybridization are indeed able to survey large genomic rearrangements and copy number changes across the entire genome, these methods lack the resolution required for detection of many mutations that are important for myeloid cancers.
0 Comments Click here to read/write comments
A New Focus on Implementing Clinical Genomics
Category: clinical genomics, NGS
Posted by
Trevor Brown on Oct 20, 2017 12:00:00 AM
For many years, next-generation sequencing (NGS) made headlines with researchers promising unprecedented breakthroughs in medical diagnostics. But the clinical impact was always explained as being a few years and more large-scale studies away from reality. In 2010, forward-thinking academics forecasted whole-genome sequencing in a matter of hours for only $30 (right around that same time, a Stanford researcher sequenced his own genome for less than $50,000 – a record low at that point).
0 Comments Click here to read/write comments
This is the Number One Risk to Your Clinical Sequencing Assay
Category: bioinformatics, NGS
Posted by
Trevor Brown on Oct 17, 2017 12:00:00 AM
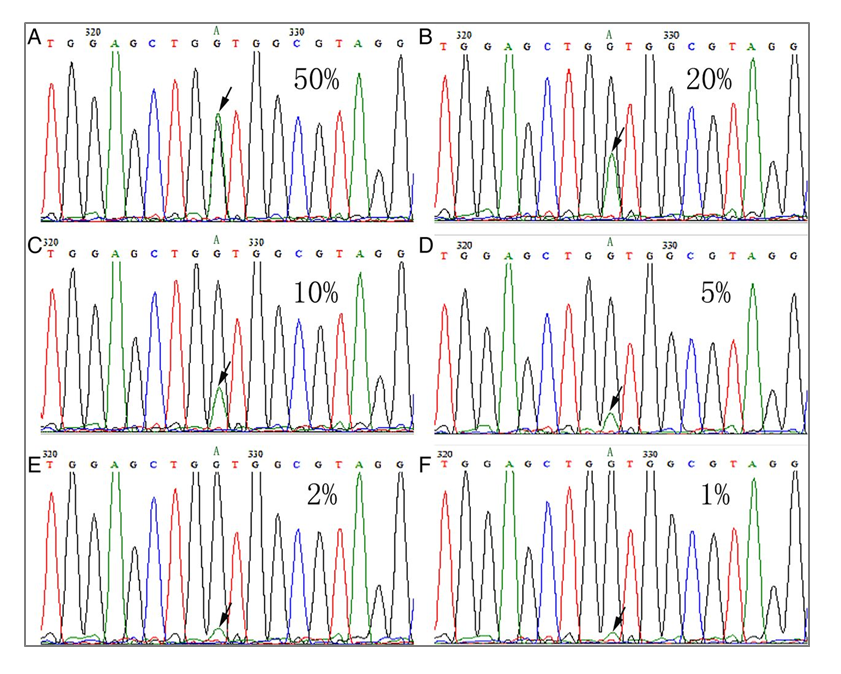
If you’re relying on remnant patient samples to tell you how well your lab's bioinformatics pipeline can call clinically important variants, you might be missing more than you realize. In our experience, the bioinformatics pipeline can be the weakest link in assay development for many labs. Just because a variant is sequenced correctly doesn’t always mean that it will be called. And false-positives are just as bad. Sometimes it’s an issue of allele frequency. For example, we’ve seen cases where labs could detect certain mutations at 10% allele frequency, but as soon as the frequency dropped to 7%, they stopped detecting it. Other cases are caused by the complexity of the variant. For example, even at low allele frequencies, a lab may pick up relatively easy-to-detect single-nucleotide variants (SNVs) but can have problems with insertion/deletion (INDEL) calling errors. In both examples, the mutations aren’t missed because of sequencing or library preparation problems. As we’ve witnessed time and time again, when labs optimize their bioinformatics pipelines, they start picking up the low-frequency and difficult-to-detect variants again. The catch is, you first have to know you’re missing something. In assay development, what you don’t know can seriously weaken your test.
0 Comments Click here to read/write comments
Is Your NGS-Based Assay on the Right TRK?
Category: qc management, QC Management Software, NGS, RNA fusion, reference materials
Posted by
Trevor Brown on Oct 9, 2017 12:00:00 AM
Despite the absence of clear guidelines or firmly established best practices, next-generation sequencing (NGS) assays are becoming the method of choice for gene fusion detection. This is significant because, although some of the cancers that contain fusion RNAs are rare, they’re now treatable thanks to new targeted therapies. If your assay can detect fusion RNAs, it can help profile tumors for important diagnostic, prognostic, and therapeutic targets, which can lead to improved patient outcomes. The old FISH method limited you to one type of fusion variant at a time; it was effective, but also slow and cumbersome. With the latest NGS techniques, detecting fusion RNAs is more efficient than ever. It’s more sensitive and can detect multiple fusions in the same assay. Nevertheless, it’s still challenging because of the complex workflows and the need to rigorously ensure performance across all fusion variants. From extraction, to library prep, to sequencing, to the bioinformatics pipeline, there are countless points where something could go wrong.
0 Comments Click here to read/write comments
Does your NGS lab struggle with quality control? [Free Guide]
Category: QC Challenges, NGS
Posted by
Meagan Gregoire on Aug 11, 2017 12:00:00 AM
As clinical genomics diagnostics continues to evolve, the primary challenge for laboratories has shifted from data acquisition, to ensuring their NGS tests are safe and effective. Whether they are developing an assay for circulating tumor DNA, validating a test to predict an antiviral therapy response, or are in production to provide parents with life changing health information, the underlining goal remains the same: your results must be accurate, precise, and consistent. High-quality, highly-multiplexed reference materials and the effective use of quality control metrics for monitoring the health of your assays are two solutions to help achieve those goals.
0 Comments Click here to read/write comments
Dr. Andrea Ferreira-Gonzalez on the Seven Benefits of Clinical Genomics Universal Standardization
Category: clinical genomics, NGS
Posted by
Dale Yuzuki on Jun 22, 2017 12:00:00 AM
As a 25-year veteran of clinical molecular diagnostics, Dr. Andrea Ferreira-Gonzalez has seen many changes in genetic technologies used in the testing laboratory. With the advent of personalized medicine and using multi-gene NGS panels as a laboratory-developed test, Dr. Ferreira-Gonzalez and other experts have agreed to lend their expertise to the design of SeraCare’s reference materials. She and other groups have participated in an interlaboratory test of standardized reference materials for detecting cancer somatic mutations, with results that will be published in the coming months.
0 Comments Click here to read/write comments
Dr. Andrea Ferreira-Gonzalez on the Seven Benefits of Clinical Genomics Universal Standardization
Category: clinical genomics, laboratory training, NGS, reference materials
Posted by
Dale Yuzuki on Jun 22, 2017 12:00:00 AM
As a 25-year veteran of clinical molecular diagnostics, Dr. Andrea Ferreira-Gonzalez has seen many changes in genetic technologies used in the testing laboratory. With the advent of personalized medicine and using multi-gene NGS panels as a laboratory-developed test, Dr. Ferreira-Gonzalez and other experts have agreed to lend their expertise to the design of SeraCare’s reference materials. She and other groups have participated in an interlaboratory test of standardized reference materials for detecting cancer somatic mutations, with results that will be published in the coming months.
0 Comments Click here to read/write comments
Real-World Needs for Genetic Testing Reference Materials
Category: NGS, reference materials
Posted by
Matt Ryder on Jun 1, 2017 12:00:00 AM
“Happy families are all alike; every unhappy family is unhappy in its own way.” I have always thought the opening of Anna Karenina applies for many things beyond familial harmony (or lack thereof). Certainly, in the world of molecular genetic diagnostics, conclusive results are usually obtained for most patients; however, there are times when a final result is more elusive than conclusive. When this occurs, it may seem as though no two challenges are ever the same. The following are real examples – presented in general terms for patient and institutional confidentiality – of difficult, unanticipated, and even bizarre cases I encountered during my time in clinical testing for predisposition to hereditary disease. Each of these situations required extraordinary effort, dedicated time, and additional resources for resolution. At the end of each day, satisfaction came from knowing that another problem solved was another patient helped in making life-altering medical management decisions.
0 Comments Click here to read/write comments