Choose your Article Focus | NGS | Molecular & Serology
This is the Number One Risk to Your Clinical Sequencing Assay
Category: bioinformatics, NGS
Posted by
Trevor Brown on Oct 17, 2017 12:00:00 AM
If you’re relying on remnant patient samples to tell you how well your lab's bioinformatics pipeline can call clinically important variants, you might be missing more than you realize. In our experience, the bioinformatics pipeline can be the weakest link in assay development for many labs. Just because a variant is sequenced correctly doesn’t always mean that it will be called. And false-positives are just as bad. Sometimes it’s an issue of allele frequency. For example, we’ve seen cases where labs could detect certain mutations at 10% allele frequency, but as soon as the frequency dropped to 7%, they stopped detecting it. Other cases are caused by the complexity of the variant. For example, even at low allele frequencies, a lab may pick up relatively easy-to-detect single-nucleotide variants (SNVs) but can have problems with insertion/deletion (INDEL) calling errors. In both examples, the mutations aren’t missed because of sequencing or library preparation problems. As we’ve witnessed time and time again, when labs optimize their bioinformatics pipelines, they start picking up the low-frequency and difficult-to-detect variants again. The catch is, you first have to know you’re missing something. In assay development, what you don’t know can seriously weaken your test.
0 Comments Click here to read/write comments
Is Your NGS-Based Assay on the Right TRK?
Category: qc management, QC Management Software, NGS, RNA fusion, reference materials
Posted by
Trevor Brown on Oct 9, 2017 12:00:00 AM
Despite the absence of clear guidelines or firmly established best practices, next-generation sequencing (NGS) assays are becoming the method of choice for gene fusion detection. This is significant because, although some of the cancers that contain fusion RNAs are rare, they’re now treatable thanks to new targeted therapies. If your assay can detect fusion RNAs, it can help profile tumors for important diagnostic, prognostic, and therapeutic targets, which can lead to improved patient outcomes. The old FISH method limited you to one type of fusion variant at a time; it was effective, but also slow and cumbersome. With the latest NGS techniques, detecting fusion RNAs is more efficient than ever. It’s more sensitive and can detect multiple fusions in the same assay. Nevertheless, it’s still challenging because of the complex workflows and the need to rigorously ensure performance across all fusion variants. From extraction, to library prep, to sequencing, to the bioinformatics pipeline, there are countless points where something could go wrong.
0 Comments Click here to read/write comments
Expert opinions on LDT transparency and standardization
Category: clinical genomics, LDT
Posted by
Sam Blier on Oct 6, 2017 12:00:00 AM
Laboratory-developed tests (LDTs) have proliferated in the absence of clear guidelines and regulations. So how can laboratorians, physicians, and patients be assured of the quality of the diagnostic result? A panel of clinical genomics experts (Girish Putcha, MD, PhD, Director of Laboratory Science, Palmetto GBA; Roger Klein, MD, Principal, JD Consulting; Elaine Lyon, PhD, Medical Director, Molecular Genetics and Genomics, ARUP Laboratories; and Russell Garlick, PhD, CSO, SeraCare) delved into this topic during the audience Q&A session of a recent webinar hosted by GenomeWeb (you can download a full report on the entire series here).
0 Comments Click here to read/write comments
You can’t validate everything… can you?
Category: clinical genomics, QC data management
Posted by
Sam Blier on Sep 21, 2017 12:00:00 AM
“Is there a limit on the type of mutations we would need to validate given that the cost per validation is high?” This question was recently posted to a panel of clinical genomics experts that we convened for a webinar hosted by GenomeWeb (you can download a full report on the entire series here). Panelists Girish Putcha, MD, PhD, Director of Laboratory Science, Palmetto GBA; Roger Klein, MD, Principal, JD Consulting; and Elaine Lyon, PhD, Medical Director, Molecular Genetics and Genomics, ARUP Laboratories, weighed in with thoughtful responses to a query that’s on the minds of many clinical laboratorians.
0 Comments Click here to read/write comments
Rapid and Early detection of Listeria to prevent food poisoning
Category: listeria
Posted by
Vidya Murthy on Sep 12, 2017 12:00:00 AM
September is National Food Safety Education Month. This is also a time to return to school and learning, so it is a good time to learn about food safety in ready-to-eat and perishable foods often packed in school lunches. Listeria is one of the most common foodborne bacteria and is found in a wide range of foods; from ready-to-eat to produce and meat. The latest draft guidance released by the U.S. Food and Drug Administration, “Control of Listeria monocytogenes in Ready-To-Eat Foods,” supports ongoing efforts by industry and government agencies to reduce the risk of Listeria monocytogenes in ready-to-eat foods (1). The guidance includes recommendations for controls involving personnel, cleaning and maintenance of equipment, and sanitation, as well as for treatments that kill Listeria and prevent it from growing during storage of food between production and consumption. The guidance emphasizes the importance of routine testing for Listeria. Therefore, it is important to learn about the methods and reagents that can be used to detect Listeria to prevent foodborne outbreaks.
0 Comments Click here to read/write comments
Accelerating Liquid Biopsy Assay Development
Category: liquid biopsy, Assay Development, ctDNA, reference materials
Posted by
Sam Blier on Sep 8, 2017 12:00:00 AM
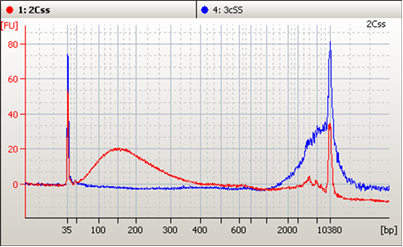
At the 2017 Next Generation Dx Summit in Washington, DC, our CSO, Russell Garlick, PhD, presented a workshop on accelerating liquid biopsy assay development. He has worked closely with a variety of groups in the liquid biopsy space that are developing and validating circulating tumor DNA (ctDNA) assays. He highlighted some common challenges facing the field, and explained how SeraCare has been using these collaborations to develop QC tools specifically for ensuring the robustness of these cutting-edge tests.
0 Comments Click here to read/write comments
Does your NGS lab struggle with quality control? [Free Guide]
Category: QC Challenges, NGS
Posted by
Meagan Gregoire on Aug 11, 2017 12:00:00 AM
As clinical genomics diagnostics continues to evolve, the primary challenge for laboratories has shifted from data acquisition, to ensuring their NGS tests are safe and effective. Whether they are developing an assay for circulating tumor DNA, validating a test to predict an antiviral therapy response, or are in production to provide parents with life changing health information, the underlining goal remains the same: your results must be accurate, precise, and consistent. High-quality, highly-multiplexed reference materials and the effective use of quality control metrics for monitoring the health of your assays are two solutions to help achieve those goals.
0 Comments Click here to read/write comments
Three liquid biopsy problems solved by the most patient-like ctDNA reference materials
Category: cfDNA, ctDNA, reference materials
Posted by
Dale Yuzuki on Jul 24, 2017 12:00:00 AM
Clinical genomics laboratories are increasingly looking to liquid biopsy cancer assays to complement their current solid tumor assays. Compared to their solid tumor assay counterparts, circulating tumor DNA (ctDNA) assays offer a different set of challenges to consider for clinical labs. One of the most important of which, is to develop a set of reagents that are appropriately validated to determine the critical performance of the assay across many parameters. The ctDNA targets of liquid biopsy assays are typically at much lower allelic frequencies and require a robust and reproducibly designed assay to consistently detect these important variants.
0 Comments Click here to read/write comments
What specifically contributes to high levels of NGS accuracy?
Category: clinical genomics
Posted by
Dale Yuzuki on Jul 7, 2017 12:00:00 AM
Accuracy of NGS results is extremely critical for clinical genomic laboratories, when developing, validating or running an assay. At the 2017 Precision Medicine World Conference a panel discussion entitled ‘Achieving Accurate NGS Test Results’ featured Dr. Greg Tsongalis, Director of Clinical Genomics and Advanced Technology at Dartmouth Hitchcock Medical Center; Dr. Dara Aisner, Associate Professor of Pathology at University of Colorado Anschutz Medical Campus School of Medicine; and Dr. Russell Garlick, Chief Scientific Officer of SeraCare Life Sciences.
0 Comments Click here to read/write comments
Dr. Andrea Ferreira-Gonzalez on the Seven Benefits of Clinical Genomics Universal Standardization
Category: clinical genomics, NGS
Posted by
Dale Yuzuki on Jun 22, 2017 12:00:00 AM
As a 25-year veteran of clinical molecular diagnostics, Dr. Andrea Ferreira-Gonzalez has seen many changes in genetic technologies used in the testing laboratory. With the advent of personalized medicine and using multi-gene NGS panels as a laboratory-developed test, Dr. Ferreira-Gonzalez and other experts have agreed to lend their expertise to the design of SeraCare’s reference materials. She and other groups have participated in an interlaboratory test of standardized reference materials for detecting cancer somatic mutations, with results that will be published in the coming months.
0 Comments Click here to read/write comments